| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,641,780 – 8,641,873 |
| Length | 93 |
| Max. P | 0.912066 |

| Location | 8,641,780 – 8,641,873 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Shannon entropy | 0.34680 |
| G+C content | 0.41267 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.66 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
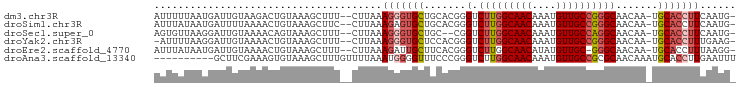
>dm3.chr3R 8641780 93 + 27905053 AUUUUUAAUGAUUGUAAGACUGUAAAGCUUU--CUUAAAGGGUGCUGCACGGGUCUUGGCAACAAAUGUUGCCGGGCAACAA-UGCACCUUCAAUG- ..........(((((((((..(.....)..)--)))).(((((((.......((((.((((((....)))))))))).....-.))))))))))).- ( -27.62, z-score = -2.10, R) >droSim1.chr3R 14741550 93 - 27517382 AUUUAUAAUGAUUUUAAAACUGUAAAGCUUC--CUUAAAGAGUGCUGCACGGGUCUUGGCAACAAAUGUUGCCGGGCAACAA-UGCACCUUCAAUG- ........(((........((((..((((((--......))).)))..)))).....((((((....))))))(((((....-))).)).)))...- ( -22.90, z-score = -1.31, R) >droSec1.super_0 7773283 91 - 21120651 AGUGUUAAGGAUUGUAAAACAGUAAAGCUUU--CUUAAAGGGUGCUGC--CGGUCUUGGCAACAAAUGUUGCCAGGCAACAA-UGCACCUUCAAUG- ....(((((((..((...........)).))--)))))(((((((...--..((((.((((((....)))))))))).....-.))))))).....- ( -27.70, z-score = -1.80, R) >droYak2.chr3R 12934459 92 + 28832112 -AUUUUAAGGAUUGUAAAACUGUAAAGCUUU--CUUAAAGGGUGCUCCACGGGUCUUGGCAACAAAUGUUGCCGGGCAACAA-UGCACCUUUGAAG- -....((((((..((...........)).))--))))((((((((.......((((.((((((....)))))))))).....-.))))))))....- ( -27.42, z-score = -2.04, R) >droEre2.scaffold_4770 4738837 92 - 17746568 AUUUAUAAUGAUUGUAAAACUGUAAAGCUUU--CUUAAAGAUUGCUUCACGGGUCUUGGCAACAUAUGUUGC-GGGCAACAA-UGCACCUUUAAGG- .((((((.....)))))).((((.((((..(--(.....))..)))).))))..(((((((((....)))))-(((((....-))).))..)))).- ( -23.70, z-score = -2.40, R) >droAna3.scaffold_13340 14178009 87 - 23697760 ----------GCUUCGAAAGUGUAAAGCUUUGUUUUAAAUGGGGUUUCCCGGGUCUUGGCAACAAAUGUUGCCGCGCAACAAAUGCACCUUGAAUUU ----------..(((((..(((((....((((((......(((....)))(.((...((((((....)))))))).)))))))))))).)))))... ( -23.10, z-score = -0.61, R) >consensus AUUUUUAAUGAUUGUAAAACUGUAAAGCUUU__CUUAAAGGGUGCUGCACGGGUCUUGGCAACAAAUGUUGCCGGGCAACAA_UGCACCUUCAAUG_ ......................................(((((((.......(.(((((((((....)))))))))).......)))))))...... (-18.77 = -19.66 + 0.89)
| Location | 8,641,780 – 8,641,873 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Shannon entropy | 0.34680 |
| G+C content | 0.41267 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -13.70 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
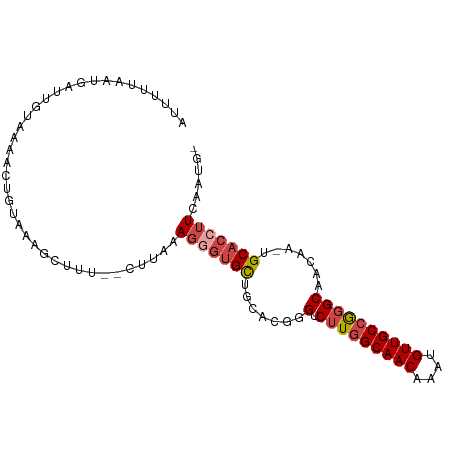
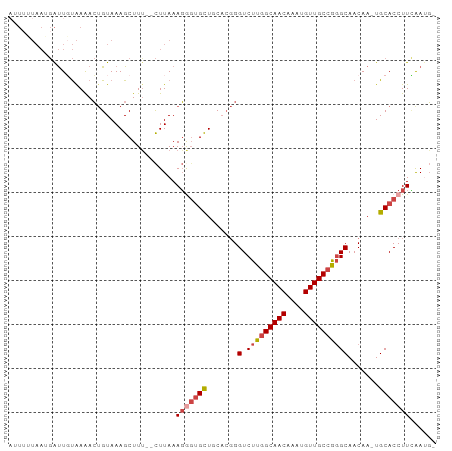
>dm3.chr3R 8641780 93 - 27905053 -CAUUGAAGGUGCA-UUGUUGCCCGGCAACAUUUGUUGCCAAGACCCGUGCAGCACCCUUUAAG--AAAGCUUUACAGUCUUACAAUCAUUAAAAAU -.((((..((.(((-....)))))((((((....))))))(((((..(((.(((..........--...))).))).))))).)))).......... ( -24.52, z-score = -2.37, R) >droSim1.chr3R 14741550 93 + 27517382 -CAUUGAAGGUGCA-UUGUUGCCCGGCAACAUUUGUUGCCAAGACCCGUGCAGCACUCUUUAAG--GAAGCUUUACAGUUUUAAAAUCAUUAUAAAU -...(((.((.(((-....)))))((((((....))))))(((((..(((.(((..(((....)--)).))).))).)))))....)))........ ( -23.50, z-score = -1.55, R) >droSec1.super_0 7773283 91 + 21120651 -CAUUGAAGGUGCA-UUGUUGCCUGGCAACAUUUGUUGCCAAGACCG--GCAGCACCCUUUAAG--AAAGCUUUACUGUUUUACAAUCCUUAACACU -..(((((((.(..-.((((((((((((((....))))))).....)--)))))))))))))).--(((((......)))))............... ( -27.70, z-score = -2.94, R) >droYak2.chr3R 12934459 92 - 28832112 -CUUCAAAGGUGCA-UUGUUGCCCGGCAACAUUUGUUGCCAAGACCCGUGGAGCACCCUUUAAG--AAAGCUUUACAGUUUUACAAUCCUUAAAAU- -.......((.(((-....)))))((((((....))))))(((((..(((((((..........--...))))))).)))))..............- ( -24.82, z-score = -2.40, R) >droEre2.scaffold_4770 4738837 92 + 17746568 -CCUUAAAGGUGCA-UUGUUGCCC-GCAACAUAUGUUGCCAAGACCCGUGAAGCAAUCUUUAAG--AAAGCUUUACAGUUUUACAAUCAUUAUAAAU -.......((.(((-....)))))-(((((....))))).(((((..(((((((..((.....)--)..))))))).)))))............... ( -22.80, z-score = -3.28, R) >droAna3.scaffold_13340 14178009 87 + 23697760 AAAUUCAAGGUGCAUUUGUUGCGCGGCAACAUUUGUUGCCAAGACCCGGGAAACCCCAUUUAAAACAAAGCUUUACACUUUCGAAGC---------- .........(((((.....)))))((((((....)))))).......(((....)))............(((((........)))))---------- ( -23.80, z-score = -1.90, R) >consensus _CAUUGAAGGUGCA_UUGUUGCCCGGCAACAUUUGUUGCCAAGACCCGUGCAGCACCCUUUAAG__AAAGCUUUACAGUUUUACAAUCAUUAAAAAU ...(((((((.(((.....)))..((((((....))))))................))))))).................................. (-13.70 = -13.98 + 0.28)

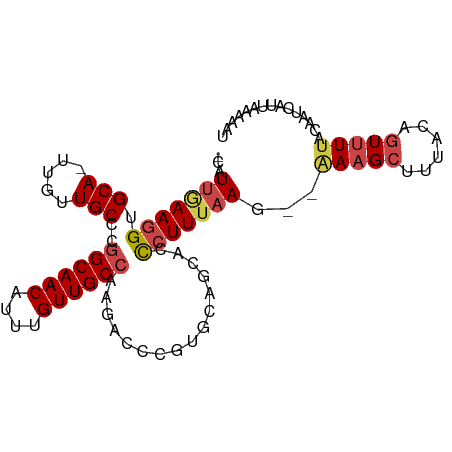
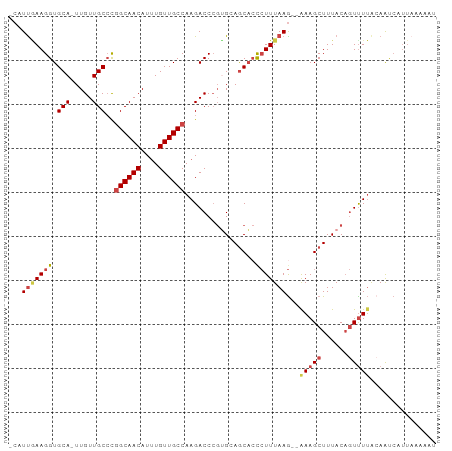
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:09 2011