
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,632,314 – 8,632,429 |
| Length | 115 |
| Max. P | 0.854012 |

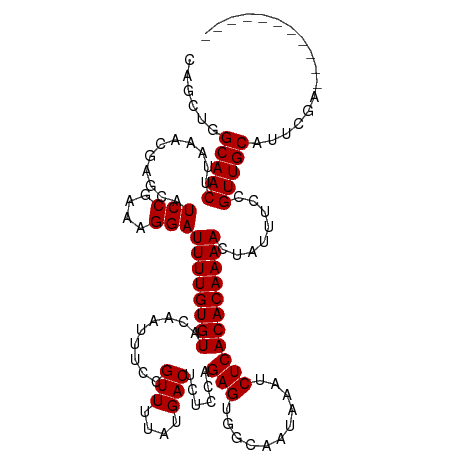
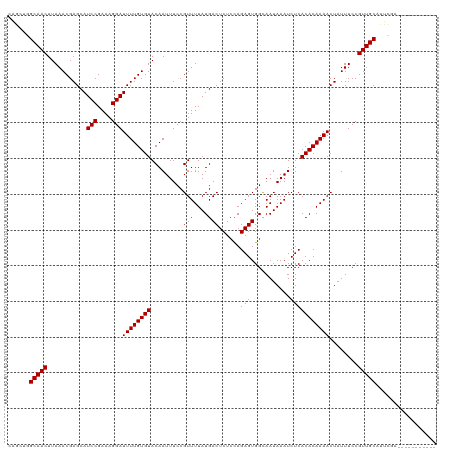
| Location | 8,632,314 – 8,632,423 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Shannon entropy | 0.11807 |
| G+C content | 0.39739 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -21.90 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 8632314 109 + 27905053 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >droSim1.chr3R 14732149 109 - 27517382 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >droSec1.super_0 7763946 109 - 21120651 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >droYak2.chr3R 12925086 109 + 28832112 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >droEre2.scaffold_4770 4729286 109 - 17746568 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >droAna3.scaffold_13340 14169265 109 - 23697760 CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))......----------- ( -21.90, z-score = -0.61, R) >dp4.chr2 19239041 120 - 30794189 CUACUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGAGCUGCUGCUGC ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........))))).....(((....))).. ( -24.40, z-score = -0.14, R) >droWil1.scaffold_181089 11351813 109 + 12369635 AGAUGGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCUAGAGUGGUAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA----------- .((...(((((...........(((....)))((((((((...((((.....((((.((((....)))).)))).))))...))))))))........)))))..))..----------- ( -27.50, z-score = -2.32, R) >droVir3.scaffold_12855 5946197 108 - 10161210 -AUGGUGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGUAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUGGA----------- -..((((((((...........(((....)))((((((((...((((.....((((.((((....)))).)))).))))...))))))))........))))))))...----------- ( -28.80, z-score = -2.65, R) >droGri2.scaffold_14906 7361264 108 - 14172833 -CGGGUGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGUAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUGGA----------- -(.((((((((...........(((....)))((((((((...((((.....((((.((((....)))).)))).))))...))))))))........)))))))).).----------- ( -30.50, z-score = -2.85, R) >droMoj3.scaffold_6540 25585821 108 - 34148556 -AUGGUGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCUAGAGUGGUAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUGGA----------- -..((((((((...........(((....)))((((((((...((((.....((((.((((....)))).)))).))))...))))))))........))))))))...----------- ( -28.80, z-score = -2.80, R) >consensus CAGCUGGCAACUUAAACGAGCAUCCGAAAGGAUUUUGUGUACAAUUUCCGUUUUAUGACUCUCCAGAGUGGCAAUAAAUCUCACACAAAACUAUUUCCGUUGCAUUCGA___________ ......(((((...........(((....)))((((((((.........(((....)))......(((...........)))))))))))........)))))................. (-21.90 = -21.90 + 0.00)

| Location | 8,632,314 – 8,632,423 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Shannon entropy | 0.11807 |
| G+C content | 0.39739 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -25.59 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8632314 109 - 27905053 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >droSim1.chr3R 14732149 109 + 27517382 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >droSec1.super_0 7763946 109 + 21120651 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >droYak2.chr3R 12925086 109 - 28832112 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >droEre2.scaffold_4770 4729286 109 + 17746568 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >droAna3.scaffold_13340 14169265 109 + 23697760 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG -----------......(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... ( -25.00, z-score = -0.55, R) >dp4.chr2 19239041 120 + 30794189 GCAGCAGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGUAG ......((((((..((((((.(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).).)))..)))..))))))...... ( -33.30, z-score = -1.63, R) >droWil1.scaffold_181089 11351813 109 - 12369635 -----------UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUAGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCCAUCU -----------..((..(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...)). ( -25.40, z-score = -1.64, R) >droVir3.scaffold_12855 5946197 108 + 10161210 -----------UCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCACCAU- -----------.....((((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..))))))....- ( -26.90, z-score = -1.99, R) >droGri2.scaffold_14906 7361264 108 + 14172833 -----------UCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCACCCG- -----------.....((((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..))))))....- ( -26.90, z-score = -1.91, R) >droMoj3.scaffold_6540 25585821 108 + 34148556 -----------UCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUAGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCACCAU- -----------.....((((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..))))))....- ( -26.90, z-score = -2.48, R) >consensus ___________UCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGAUGCUCGUUUAAGUUGCCAGCUG .................(((((..(((((((((((((((.(((((.(((..((((....))))..))).....))))).))))))))((((....))))))).))))..)))))...... (-25.59 = -25.35 + -0.23)
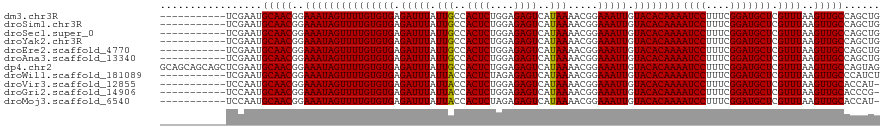


| Location | 8,632,336 – 8,632,429 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Shannon entropy | 0.05097 |
| G+C content | 0.38953 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.32 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8632336 93 - 27905053 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droSim1.chr3R 14732171 93 + 27517382 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droSec1.super_0 7763968 93 + 21120651 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droYak2.chr3R 12925108 93 - 28832112 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droEre2.scaffold_4770 4729308 93 + 17746568 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droAna3.scaffold_13340 14169287 93 + 23697760 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.90, z-score = -1.04, R) >droWil1.scaffold_181089 11351835 93 - 12369635 AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUAGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -22.20, z-score = -1.85, R) >droVir3.scaffold_12855 5946218 90 + 10161210 ---AGCUCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA ---.((......))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.50, z-score = -1.69, R) >droGri2.scaffold_14906 7361285 90 + 14172833 ---AGCUCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA ---.((......))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -21.50, z-score = -1.69, R) >droMoj3.scaffold_6540 25585842 93 + 34148556 AGCAGCUCCAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUACCACUCUAGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA .(((.......)))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). ( -22.20, z-score = -2.03, R) >consensus AGCAGCUCGAAUGCAACGGAAAUAGUUUUGUGUGAGAUUUAUUGCCACUCUGGAGAGUCAUAAAACGGAAAUUGUACACAAAAUCCUUUCGGA ....((......))..(.((((..((((((((((.(((((.(((..((((....))))..))).....))))).))))))))))..)))).). (-21.56 = -21.32 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:07 2011