
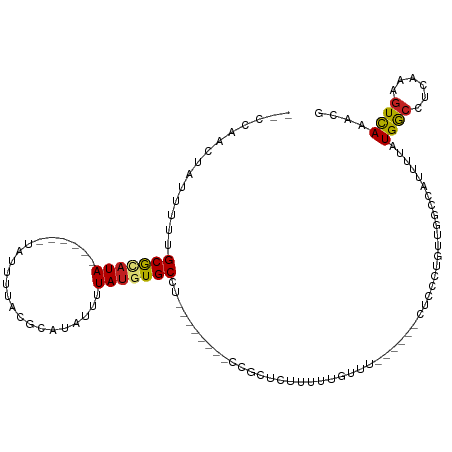
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,610,368 – 8,610,464 |
| Length | 96 |
| Max. P | 0.533158 |

| Location | 8,610,368 – 8,610,464 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.23 |
| Shannon entropy | 0.55927 |
| G+C content | 0.38302 |
| Mean single sequence MFE | -18.15 |
| Consensus MFE | -5.55 |
| Energy contribution | -5.31 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
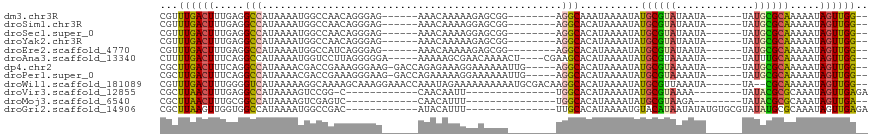
>dm3.chr3R 8610368 96 + 27905053 --CCAACUAUUUUUGCGCAUA------UAUUAUACGCAUAUUUUAUUUGCCU--------CCGCUCUUUUUGUUU------CUCCCUGUUGGCCAUUUUAUGGCCUCAAAGUCAAACG --..............(((.(------((..(((....)))..))).)))..--------...........((((------..(..((..((((((...)))))).))..)..)))). ( -14.30, z-score = -1.33, R) >droSim1.chr3R 14708872 96 - 27517382 --CCAACUAUUUUUGCGCAUA------UAUUAUACGCAUAUUUUAUGUGCCU--------CCGCUCCUUUUGUUU------CUCCCUGUUGGCCAUUUUAUGGCCUCAAAGUCAAACG --............(((((((------...(((....)))...)))))))..--------...........((((------..(..((..((((((...)))))).))..)..)))). ( -18.40, z-score = -2.57, R) >droSec1.super_0 7742251 96 - 21120651 --CCAACUAUUUUUGCGCAUA------UAUUAUACGCAUAUUUUAUGUGCCU--------CCGCUCCUUUUGUUU------CUCCCUGUUGGCCAUUUUAUGGCCUCAAAGUCAAACG --............(((((((------...(((....)))...)))))))..--------...........((((------..(..((..((((((...)))))).))..)..)))). ( -18.40, z-score = -2.57, R) >droYak2.chr3R 12902779 96 + 28832112 --CCAACUAUUUUUGCGCAUA------UAUUAUACGCAUAUUUUAUGUGCCU--------CCGCUCUUUUUGUUU------CUCCCUGUUGGCCAUUUUAUGGCCUCAAAGUCAAACG --............(((((((------...(((....)))...)))))))..--------...........((((------..(..((..((((((...)))))).))..)..)))). ( -18.40, z-score = -2.54, R) >droEre2.scaffold_4770 4706900 96 - 17746568 --CCAACUAUUUUUGCGCAUA------UAUUAUACGCAUAUUUUAUGUGCCU--------CCGCUCUUUUUGUUU------CUCCCUGAUGGCCAUUUUAUGGCCUCAAAGUCAAACG --............(((((((------...(((....)))...)))))))..--------...........((((------..(..(((.((((((...)))))))))..)..)))). ( -20.30, z-score = -3.09, R) >droAna3.scaffold_13340 14149279 101 - 23697760 --CCAACUAUUUUUGCAAAUA------UAUUUUACGCAUAUUUUAUGUGCUUCG----AGUUUUGUUCGCUUUUU-----UCCCCCUAAGGACCAUUUUAUGGCCUGAAAGUCAAAAG --......((((((((.((((------.((((...((((((...))))))...)----)))..)))).)).....-----........(((.((((...)))))))))))))...... ( -14.00, z-score = -0.19, R) >dp4.chr2 19214287 104 - 30794189 --CCAACUAUUUUUGCGCAUA------UAUUUUACGCAUAUUUUAUGUGCCU-----CAAUUUUUUCCUUUCUCUGGUC-CUUCCCUUUCGGUCGUUUUAUGGCCUGAAAGUCAAGCG --............(((((((------................)))))))..-----.........((.......))..-(((..(((((((((((...)))))).)))))..))).. ( -18.39, z-score = -1.96, R) >droPer1.super_0 8940585 104 + 11822988 --CCAACUAUUUUUGCGCAUA------UAUUUUACGCAUAUUUUAUGUGCCU-----CAAUUUUUUCCUUUUUCUGGUC-CUUCCCUUUCGGUCGUUUUAUGGCCUGAAAGUCAAGCG --............(((((((------................)))))))..-----.........((.......))..-(((..(((((((((((...)))))).)))))..))).. ( -18.39, z-score = -1.95, R) >droWil1.scaffold_181089 11329620 108 + 12369635 --CCAACUAUUUUUGCG--UA------UAUUUAACGCAUAUUUUAUGUGCCUUGUCGCAUUUUUUUUUUCUAUUUGGUUUCCUUUGCUUUUGCCUUUUUAUGACCCCAAAGUCAAACG --((((.......((((--(.------......)))))........((((......)))).............))))........((....)).......((((......)))).... ( -13.20, z-score = -1.02, R) >droVir3.scaffold_12855 5916906 82 - 10161210 UCUCAACUAUUUGCGCGUAUA--------UUUUACGCAUAUUUUAUGUGCCA---------------AAUUGUUG------------G-CCGGACUUUUAUGGCCUCAAAGUUAAGCG ............(((((((..--------...)))))...............---------------..(((..(------------(-(((........))))).)))......)). ( -18.30, z-score = -1.15, R) >droMoj3.scaffold_6540 25555266 81 - 34148556 --UCAACUAUUUGCGCGUAUA--------UCUUACGCAUAUUUUAUGUGCCA---------------AAAUGUUG------------GACUCGACUUUUAUGGCCGCAAAGUUAAGCG --..((((..(((((((((..--------...))))).........(.((((---------------(((.((((------------....)))).))).)))))))))))))..... ( -20.80, z-score = -2.02, R) >droGri2.scaffold_14906 7340267 91 - 14172833 UCUCAACUAUUUGCGCAUAUACGCACAUAUAUUAUGUACAUUUUAUGUGCAA---------------AAAUGUAU------------GUCGGCCAUUUUAUGGCCACCAACUUAAGCG .............(((......(.(((((((((.(((((((...))))))).---------------.)))))))------------)))((((((...))))))..........))) ( -24.90, z-score = -3.44, R) >consensus __CCAACUAUUUUUGCGCAUA______UAUUUUACGCAUAUUUUAUGUGCCU________CCGCUCUUUUUGUUU______CUCCCUGUUGGCCAUUUUAUGGCCUCAAAGUCAAACG ...................................((((((...))))))..................................................((((......)))).... ( -5.55 = -5.31 + -0.24)

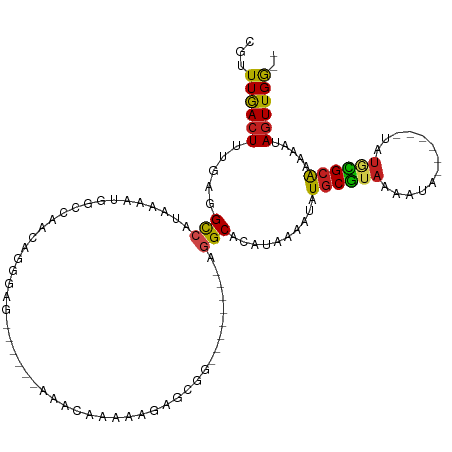
| Location | 8,610,368 – 8,610,464 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.23 |
| Shannon entropy | 0.55927 |
| G+C content | 0.38302 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -6.01 |
| Energy contribution | -5.60 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8610368 96 - 27905053 CGUUUGACUUUGAGGCCAUAAAAUGGCCAACAGGGAG------AAACAAAAAGAGCGG--------AGGCAAAUAAAAUAUGCGUAUAAUA------UAUGCGCAAAAAUAGUUGG-- .((((..(((((.((((((...))))))..)))))..------)))).......((..--------..))..........((((((((...------))))))))...........-- ( -23.80, z-score = -3.24, R) >droSim1.chr3R 14708872 96 + 27517382 CGUUUGACUUUGAGGCCAUAAAAUGGCCAACAGGGAG------AAACAAAAGGAGCGG--------AGGCACAUAAAAUAUGCGUAUAAUA------UAUGCGCAAAAAUAGUUGG-- .((((..(((((.((((((...))))))..)))))..------)))).......((..--------..))..........((((((((...------))))))))...........-- ( -23.80, z-score = -2.85, R) >droSec1.super_0 7742251 96 + 21120651 CGUUUGACUUUGAGGCCAUAAAAUGGCCAACAGGGAG------AAACAAAAGGAGCGG--------AGGCACAUAAAAUAUGCGUAUAAUA------UAUGCGCAAAAAUAGUUGG-- .((((..(((((.((((((...))))))..)))))..------)))).......((..--------..))..........((((((((...------))))))))...........-- ( -23.80, z-score = -2.85, R) >droYak2.chr3R 12902779 96 - 28832112 CGUUUGACUUUGAGGCCAUAAAAUGGCCAACAGGGAG------AAACAAAAAGAGCGG--------AGGCACAUAAAAUAUGCGUAUAAUA------UAUGCGCAAAAAUAGUUGG-- .((((..(((((.((((((...))))))..)))))..------)))).......((..--------..))..........((((((((...------))))))))...........-- ( -23.80, z-score = -3.01, R) >droEre2.scaffold_4770 4706900 96 + 17746568 CGUUUGACUUUGAGGCCAUAAAAUGGCCAUCAGGGAG------AAACAAAAAGAGCGG--------AGGCACAUAAAAUAUGCGUAUAAUA------UAUGCGCAAAAAUAGUUGG-- .((((..((((((((((((...)))))).))))))..------)))).......((..--------..))..........((((((((...------))))))))...........-- ( -27.00, z-score = -3.93, R) >droAna3.scaffold_13340 14149279 101 + 23697760 CUUUUGACUUUCAGGCCAUAAAAUGGUCCUUAGGGGGA-----AAAAAGCGAACAAAACU----CGAAGCACAUAAAAUAUGCGUAAAAUA------UAUUUGCAAAAAUAGUUGG-- (((((..((..((((((((...)))).)))..)..)).-----.)))))(.(((......----....(((.........)))(((((...------..))))).......))).)-- ( -16.40, z-score = -0.29, R) >dp4.chr2 19214287 104 + 30794189 CGCUUGACUUUCAGGCCAUAAAACGACCGAAAGGGAAG-GACCAGAGAAAGGAAAAAAUUG-----AGGCACAUAAAAUAUGCGUAAAAUA------UAUGCGCAAAAAUAGUUGG-- ...(..(((.....(((.........((....))....-..((.......)).........-----.)))..........((((((.....------..)))))).....)))..)-- ( -19.30, z-score = -1.34, R) >droPer1.super_0 8940585 104 - 11822988 CGCUUGACUUUCAGGCCAUAAAACGACCGAAAGGGAAG-GACCAGAAAAAGGAAAAAAUUG-----AGGCACAUAAAAUAUGCGUAAAAUA------UAUGCGCAAAAAUAGUUGG-- ...(..(((.....(((.........((....))....-..((.......)).........-----.)))..........((((((.....------..)))))).....)))..)-- ( -19.30, z-score = -1.45, R) >droWil1.scaffold_181089 11329620 108 - 12369635 CGUUUGACUUUGGGGUCAUAAAAAGGCAAAAGCAAAGGAAACCAAAUAGAAAAAAAAAAUGCGACAAGGCACAUAAAAUAUGCGUUAAAUA------UA--CGCAAAAAUAGUUGG-- (.((((.((((...(((.......))).)))))))).)...((((..............(((......))).........(((((......------.)--)))).......))))-- ( -18.20, z-score = -1.91, R) >droVir3.scaffold_12855 5916906 82 + 10161210 CGCUUAACUUUGAGGCCAUAAAAGUCCGG-C------------CAACAAUU---------------UGGCACAUAAAAUAUGCGUAAAA--------UAUACGCGCAAAUAGUUGAGA ..(((((((.....((............(-(------------(((....)---------------))))...........(((((...--------..)))))))....))))))). ( -20.70, z-score = -2.21, R) >droMoj3.scaffold_6540 25555266 81 + 34148556 CGCUUAACUUUGCGGCCAUAAAAGUCGAGUC------------CAACAUUU---------------UGGCACAUAAAAUAUGCGUAAGA--------UAUACGCGCAAAUAGUUGA-- ...((((((((((.((((.(((.((.(....------------).)).)))---------------))))...........(((((...--------..)))))))))..))))))-- ( -18.10, z-score = -1.02, R) >droGri2.scaffold_14906 7340267 91 + 14172833 CGCUUAAGUUGGUGGCCAUAAAAUGGCCGAC------------AUACAUUU---------------UUGCACAUAAAAUGUACAUAAUAUAUGUGCGUAUAUGCGCAAAUAGUUGAGA ..(((((((((.(((((((...))))))).)------------).))...(---------------((((.((((..(((((((((...))))))))).)))).)))))...))))). ( -28.50, z-score = -2.90, R) >consensus CGUUUGACUUUGAGGCCAUAAAAUGGCCAACAGGGAG______AAACAAAAAGAGCGG________AGGCACAUAAAAUAUGCGUAAAAUA______UAUGCGCAAAAAUAGUUGG__ ...((((((.....(((..................................................)))..........((((((.............)))))).....)))))).. ( -6.01 = -5.60 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:10:02 2011