| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,584,783 – 8,584,893 |
| Length | 110 |
| Max. P | 0.650191 |

| Location | 8,584,783 – 8,584,893 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.83 |
| Shannon entropy | 0.52858 |
| G+C content | 0.52497 |
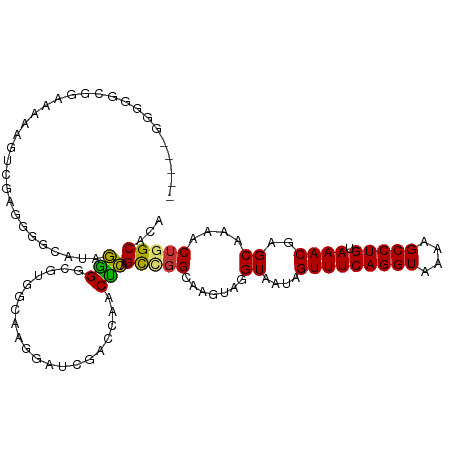
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
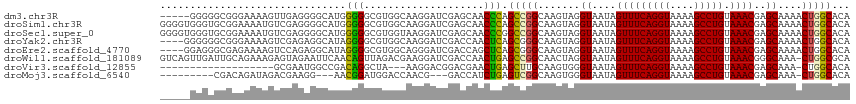
>dm3.chr3R 8584783 110 + 27905053 -----GGGGGCGGGAAAAGUUGAGGGGCAUGGGGGCGUGGCAAGGAUCGAGCAACCCAGCCGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA -----...((((((.....((((..(.((((....)))).).....))))....))).))).((.(((..((....(((((((((....))))).))))..))...))).))... ( -29.50, z-score = 0.15, R) >droSim1.chr3R 14681573 115 - 27517382 GGGGUGGGUGCGGAAAAUGUCGAGGGGCAUGGGGGCGUGGCAAGGAUCGAGCAACCCAGCCGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA .((.(((((((........((((..(.((((....)))).).....)))))).))))).)).((.(((..((....(((((((((....))))).))))..))...))).))... ( -35.00, z-score = -0.82, R) >droSec1.super_0 7716703 115 - 21120651 GGGGUGGGUGCGGAAAAUGUCGAGGGGCAUGGGGGCGUGGUAAGGAUCGAGCAACCCGGCCGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA .((.(((((.......(((((....)))))....((.((((....)))).)).))))).)).((.(((..((....(((((((((....))))).))))..))...))).))... ( -32.90, z-score = -0.25, R) >droYak2.chr3R 12875293 111 + 28832112 ----GGGGGGCGGGAAAAGUCGAGAGGCAUAGGGGCGUGGCAAGGAUCGACCAACUCAGCGGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA ----.....((.......(((....)))...(((...((((.......).))).))).))..((.(((..((....(((((((((....))))).))))..))...))).))... ( -25.60, z-score = 0.23, R) >droEre2.scaffold_4770 4681243 111 - 17746568 ----GGAGGGCGAGAAAAGUCCAGAGGCAUAGGGGCGUGGCAGGGAUCGACCAGCUCAGCGGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA ----...((((.......))))....((......))(((.(((((.....)).((((.((......))........(((((((((....))))).))))))))....))).))). ( -33.30, z-score = -1.89, R) >droWil1.scaffold_181089 11303706 114 + 12369635 GUCAGUUGAUUGCAGAAAGAGUAGAAUUCAACAGUUAGACGAAGGAUCGACCAACUGAGCCGGCAACUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGGGCAAA-CUGGCGCA (((((((........................(((((.(.(((....))).).))))).((((....).........(((((((((....))))).)))).))).))-)))))... ( -31.30, z-score = -1.81, R) >droVir3.scaffold_12855 5886720 92 - 10161210 -------------------GCGAAUGGCCGACAGGCUA---AAGGACGGACGAACUGAGCUUGCAAGUGGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAA-CUGGCACA -------------------((...(((((....)))))---.....(((.....))).)).(((.(((..((....(((((((((....))))).))))..))..)-)).))).. ( -25.80, z-score = -1.07, R) >droMoj3.scaffold_6540 25519762 99 - 34148556 ---------CGACAGAUAGACGAAGG---AACGGAUGGACCAACG---GACCAUCUGAGUCGGCAAGUGGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAA-CUGGCACA ---------((((.......(....)---..(((((((.((...)---).))))))).))))((.(((..((....(((((((((....))))).))))..))..)-)).))... ( -32.20, z-score = -3.45, R) >consensus _____GGGGGCGGAAAAAGUCGAGGGGCAUAGGGGCGUGGCAAGGAUCGACCAACUCAGCCGGCAAGUAGGUAAUAGUUUCAGGUAAAAGCCUGUAAACGAGCAAAACUGGCACA ...............................(((....................))).(((((.......((....(((((((((....))))).))))..))....)))))... (-16.71 = -16.01 + -0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:57 2011