| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,562,435 – 8,562,538 |
| Length | 103 |
| Max. P | 0.971688 |

| Location | 8,562,435 – 8,562,538 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.44919 |
| G+C content | 0.41198 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
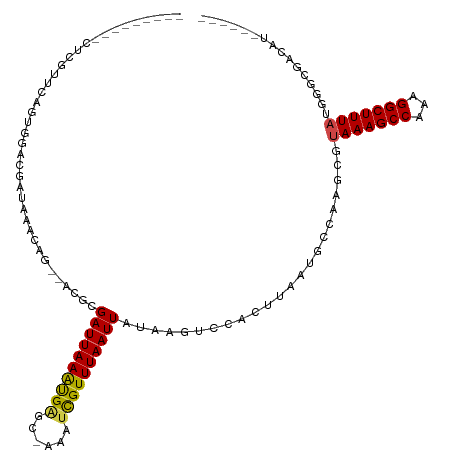
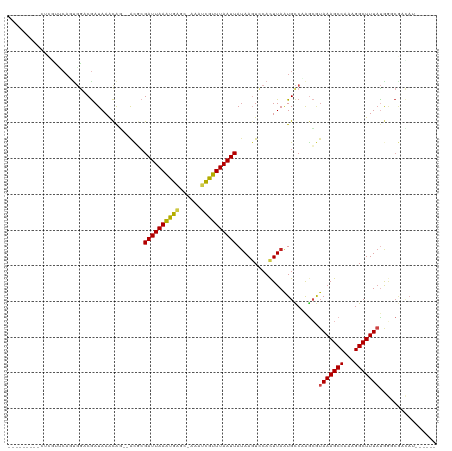
>dm3.chr3R 8562435 103 + 27905053 --------GCUCGUUCAGUGGACGAUAAACGG--ACGCGAUUAAAUGAGC-AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCCAGCGUAAAGCCAAAGGCUUUAUGGGCGACAU------ --------..(((((.....))))).....((--((..((((((((((..-...))))))))))....))))......((((((...(((((((...)))))))))))))....------ ( -35.10, z-score = -4.07, R) >droSim1.chr3R 14658089 103 - 27517382 --------GCUCGUUCAGUGGACGAUAAACGG--ACGCGAUUAAAUGAGC-AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCCAGCGUAAAGCCAAAGGCUUUAUGGGCGACAU------ --------..(((((.....))))).....((--((..((((((((((..-...))))))))))....))))......((((((...(((((((...)))))))))))))....------ ( -35.10, z-score = -4.07, R) >droSec1.super_0 7697556 103 - 21120651 --------GCUCGUUCAGUGGACGAUAAACGG--ACGCGAUUAAAUGAGC-AAAUUGUUUAAUUAUAAGUCCACUUAAUGCCCAGCGUAAAGCCAAAGGCUUUAUGGGCGACAU------ --------(((((((..(((..((.....)).--.))).....)))))))-....((((......((((....))))..(((((...(((((((...)))))))))))))))).------ ( -32.60, z-score = -3.30, R) >droYak2.chr3R 12850335 103 + 28832112 --------GCUCGUUCAGUGGACGAUAAACGG--ACGCGAUUAAAUGAGC-AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCCAGCGUAAAGCCAAAGGCUUUAUGGGCGACAU------ --------..(((((.....))))).....((--((..((((((((((..-...))))))))))....))))......((((((...(((((((...)))))))))))))....------ ( -35.10, z-score = -4.07, R) >droEre2.scaffold_4770 4660880 103 - 17746568 --------GCUCGUUCAGUGGACGAUAAACGG--ACGCGAUUAAAUGAGC-AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCCAGCGUAAAGCCAAAGGCUUUAUGGGCGCCAU------ --------(((((((.....))))).....((--((..((((((((((..-...))))))))))....)))).......(((((...(((((((...))))))))))))))...------ ( -35.50, z-score = -3.81, R) >droAna3.scaffold_13340 14108983 103 - 23697760 --------ACUCGUCCAUCGAACGAUAAACAG--ACGCGAUUAAAUGAGC-AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCAAGCGUAAAGCCAAAGGCUUUAUGAGCGCUCU------ --------....((..(((....)))..))((--((((((((((((((..-...)))))))))).((((....))))........(((((((((...))))))))).))).)))------ ( -26.30, z-score = -2.69, R) >dp4.chr2 19168722 104 - 30794189 -------------UGCCCUGGCCGAUAAACAG--GCGCGAUUAAAUGAGA-AAAUCGUUUAAUUAUAAUUCCACUUAAUGCCAGGCGUAAAGCCGAAGGCUUUAUAAACGACAUUUGCUG -------------.(((((((((........)--))..((((((((((..-...)))))))))).................)))).((((((((...))))))))...........)).. ( -26.70, z-score = -1.92, R) >droPer1.super_0 8894057 104 + 11822988 -------------UGCAGUGGCCGAUAAACAG--GCGCGAUUAAAUGAGA-AAAUCGUUUAAUUAUAAUUCCACUUAAUGCCAGGCGUAAAGCCGAAGGCUUUAUAGACGACAUUUGCUG -------------.(((((((((........)--))..((((((((((..-...)))))))))).......))).(((.(((.(((.....)))...))).)))...........))).. ( -27.70, z-score = -1.98, R) >droWil1.scaffold_181089 11286395 93 + 12369635 -------------UUCAAUGAACAAUAAAUACGCACACGAUUAAGCGAGC-AAAUCGUUUAAUUACAAGUCCACUUAAUGCCAAAGCUAAAGCCAAAGGCUUUCUAU------------- -------------...................(((...((((((((((..-...))))))))))..(((....)))..))).......((((((...))))))....------------- ( -16.20, z-score = -1.90, R) >droMoj3.scaffold_6540 8655323 102 + 34148556 ----------AUGCGCAAACGACGAUAAACAC--AUCCGAUUAAAUGGGCCAAAGCGUUUAAUUAUAAGUUCACUUAAUGUCAAACAGAAAGCCAAAGGCUUUACGGCCAACAG------ ----------.((.((....((((........--....(((((((((........))))))))).((((....)))).))))......((((((...))))))...))))....------ ( -18.90, z-score = -0.83, R) >droVir3.scaffold_12855 5863454 95 - 10161210 ----------------AUUCGACGAUAAACAC--ACCCGAUUAAAUGGGC-AGAGCGUUUAAUUAUAAGUUCACUUAAUGUCAAGCAUAAAGCCAAAGGCUUUACGGCCAACAA------ ----------------....((((.(((((.(--.((((......)))).-...).)))))....((((....)))).))))..((.(((((((...)))))))..))......------ ( -18.80, z-score = -1.39, R) >droGri2.scaffold_14906 7296729 111 - 14172833 ACUCGUUGCCGUUCUCAUUGGUCGAUAAGCAU--AUCGGAUUAAAUGGGA-AAAUCGUUUAAUUAUAAGUUCAUUUAGUGCCGAGGAUAAAGCCAAAGGCUUUAUGACCACAAA------ ..................(((((.........--.((((((((((((((.-..................)))))))))).))))..((((((((...)))))))))))))....------ ( -25.91, z-score = -1.20, R) >consensus _________CUCGUUCAGUGGACGAUAAACAG__ACGCGAUUAAAUGAGC_AAAUCGUUUAAUUAUAAGUCCACUUAAUGCCAAGCGUAAAGCCAAAGGCUUUAUGGGCGACAU______ ......................................((((((((((......)))))))))).......................(((((((...)))))))................ (-13.16 = -13.21 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:54 2011