| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,521,923 – 8,522,036 |
| Length | 113 |
| Max. P | 0.535858 |

| Location | 8,521,923 – 8,522,036 |
|---|---|
| Length | 113 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Shannon entropy | 0.51083 |
| G+C content | 0.46276 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.06 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
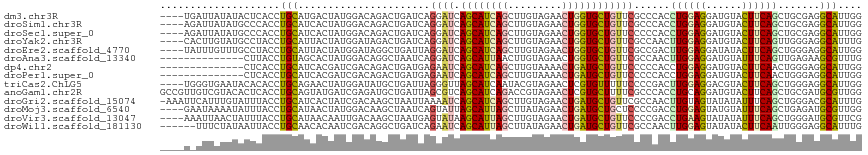
>dm3.chr3R 8521923 113 - 27905053 ----UGAUUAUAUACUCACCUGCAUGACUAUGGACAGACUGAUCAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCGCCCACCUGGAGGAUGUACUUCAGCUGCGAGGCAUUGG ----((((((((((.(((......))).)))).......)))))).((.(((((((((.........))))))))).))(((((.((((((......)))))).))...)))..... ( -30.71, z-score = 0.51, R) >droSim1.chr3R 14616338 113 + 27517382 ----AGAUUAUAUGCCCACCUGCAUCACUAUGGACAGACUGAUCAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCGCCCACCUGGAGGAUGUACUUCAGCUGCGAGGCAUUGG ----.......(((((..((((.((((((......))..))))))))..(((((((((.........))))))))).((((....((((((......))))))..)))))))))... ( -36.70, z-score = -0.76, R) >droSec1.super_0 7657563 113 + 21120651 ----AGAUUAUAUGCCCACCUGCAUCACUAUGGACAGACUGAUCAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCCCCCACCUGGAGGAUGUACUUCAGCUGCGAGGCAUUGG ----.......(((((....((((......(((...((....)).(((.(((((((((.........))))))))).))).))).((((((......)))))).)))).)))))... ( -33.50, z-score = 0.03, R) >droYak2.chr3R 12809160 113 - 28832112 ----CACUUGUAUGCCUACCUGCAUUACUAUGGAUAGACUGAUCAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCGCCAACUUGGAGGAUGUACUUCAGUUGGGAGGCAUUUG ----.......((((((.((((.(((((((....)))..))))))))..(((((((((.........)))))))))....((((((.((((......)))))))))).))))))... ( -37.80, z-score = -1.90, R) >droEre2.scaffold_4770 4617282 113 + 17746568 ----UAUUUGUUUGCCUACCUGCAUUACUAUGGAUAGGCUGAUUAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCGCCGACUUGGAGGAUAUACUUCAGCUGGGAGGCAUUGG ----.....(((.(((((((...........)).))))).)))...((.(((((((((.........))))))))).))(((..(((((((......)))))...))..)))..... ( -35.50, z-score = -1.18, R) >droAna3.scaffold_13340 17043197 103 + 23697760 --------------CUUACCUGUAGCACUAUGGACAGGCUAAUCAGGAUCAGCAUUAACUUGUAGAACUGGUGCUGUUCGCCAACUUGGAGGAUGUAUUUCAGUUGAGAAGCGUUUG --------------....((((((((.((......))))))..))))..((((((((...........))))))))(((..(((((.((((......))))))))).)))....... ( -24.00, z-score = 0.66, R) >dp4.chr2 20136129 103 + 30794189 --------------CUCACCUGCAUCACGAUCGACAGACUGAUGAGAAUCAGCAUCAGCUUGUAAAACUGAUGCUGUUCCCCCACCUGGAGGAUGUACUUCAACUGGGAGGCAUUGG --------------....(((.(((((.(.((....)))))))).(((.(((((((((.((....)))))))))))))).((((..(((((......)))))..)))))))...... ( -33.60, z-score = -2.06, R) >droPer1.super_0 9822008 103 - 11822988 --------------CUCACCUGCAUCACGAUCGACAGACUGAUGAGAAUCAGCAUCAGCUUGUAAAACUGAUGCUGUUCCCCCACCUGGAGGAUGUACUUCAACUGGGAGGCAUUGG --------------....(((.(((((.(.((....)))))))).(((.(((((((((.((....)))))))))))))).((((..(((((......)))))..)))))))...... ( -33.60, z-score = -2.06, R) >triCas2.ChLG5 4991743 113 + 18847211 ----UGGGUGAAUACACACCUGCAGAACUAUGGAUAUGCUGAUUAGGGUUAGCAUCAAUACGUAGAACUCGUGUUUUUCCCCGACUUGGAGGACGUACUUCAGCUGGGAGGCAUUGG ----.(((((......)))))......(((((...((((((((....)))))))).....)))))..((.((((..(((((.(.((.((((......))))))).))))))))).)) ( -32.30, z-score = -0.83, R) >anoGam1.chr2R 16080022 117 - 62725911 GCCGUUGUCGUACACUCACCUGCAGUAUGAUCGAGAUGCUGAUUAGCGUCAGCAUCAGACCGUAGAACUCGUGCUUUUCGCCCACCUGCAGGAUGUACUUCAGCUGCGAUGCGUUGG ((.((((..(((((....(((((((((((.((..(((((((((....))))))))).)).))))......(.((.....)).)..))))))).)))))..)))).)).......... ( -41.80, z-score = -2.03, R) >droGri2.scaffold_15074 1560286 116 - 7742996 -AAAUUCAUUUGUAUUUACCUGCAUCACUAUCGACAAGCUAAUUAAAAUCAGCAUCAGCUUGUAGAACUGAUGCUGUUCGCCAACUUGUAGUAUAUAUUUCAGCUGGGACGCAUUUG -.....((..((..(((..(((...........(((((...........(((((((((.........)))))))))........)))))...........)))...)))..))..)) ( -21.56, z-score = 0.14, R) >droMoj3.scaffold_6540 28128241 113 - 34148556 ----GAAUAAAAUAUUUACCUGCAUAACUAUGGACAAGCUAAUCAGUAUUAGCAUUAGCUUAUAGAACUGAUGCUGCUCCCCGACCUGGAGUAUGUAUUUCAGCUGAGAUGCGUUGG ----..............((...........)).(((((...(((((...((((((((.........))))))))(((((.......)))))..........)))))...)).))). ( -24.70, z-score = -0.45, R) >droVir3.scaffold_13047 11159846 113 + 19223366 ----AAAUUAACUAUUUACCUGCAUAACAAUUGACAAGCUAAUGAGUAUAAGCAUUAGCUUGUAGAACUGAUGCUGUUCCCCGACCUGAAGUAUAUAUUUCAGCUGGGAUGCGUUCG ----.................((((........(((((((((((........))))))))))).((((.......))))((((..(((((((....))))))).))))))))..... ( -31.80, z-score = -3.65, R) >droWil1.scaffold_181130 2441705 111 - 16660200 ------UUUCUAUAAUUACCUGCAACACAAUCGACAGGCUGAUCAGAAUCAGCAUUAGCUUAUAGAACUGAUGCUGUUCGCCAACUUGGAGUAUAUACUUCAAUUGGGAGGCAUUUG ------.((((((((....((((.........).)))((((((....))))))......))))))))..((((((..((.((((.(((((((....))))))))))))))))))).. ( -30.20, z-score = -2.72, R) >consensus ____U_AUUAUAUACUCACCUGCAUCACUAUGGACAGGCUGAUCAGGAUCAGCAUCAGCUUGUAGAACUGGUGCUGUUCGCCCACCUGGAGGAUGUACUUCAGCUGGGAGGCAUUGG ....................(((......................((((.((((((((.........))))))))))))......((((((......)))))).......))).... (-16.49 = -16.06 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:49 2011