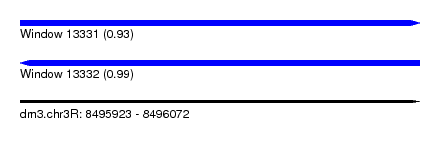
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,495,923 – 8,496,072 |
| Length | 149 |
| Max. P | 0.993126 |

| Location | 8,495,923 – 8,496,072 |
|---|---|
| Length | 149 |
| Sequences | 12 |
| Columns | 149 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.44401 |
| G+C content | 0.65607 |
| Mean single sequence MFE | -63.19 |
| Consensus MFE | -34.97 |
| Energy contribution | -34.68 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
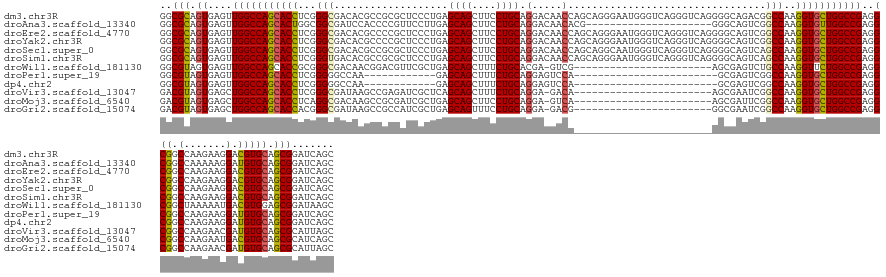
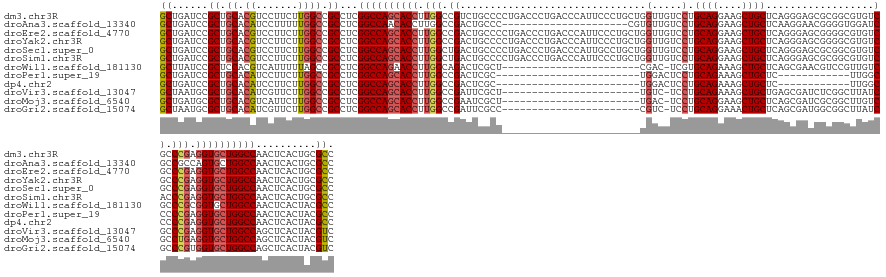
>dm3.chr3R 8495923 149 + 27905053 GGCGCAGUGAGUUGGCCAGCACCUCGGGCGACACGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAGACGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((......(((((((((((((.((.((....(((.(...((((((.(...(((((((.((....)).)))))))...).))))))...).)))...)).)).))))))))))))).(((.((.....)).)))...)))....... ( -75.60, z-score = -2.56, R) >droAna3.scaffold_13340 17017499 128 - 23697760 GGCGCAGUGAGUUGGCCAGCACUGGCGGCGAUCCACCCCGUUCCUUGAGCAGCUUCCUGCAGGACAACACG---------------------GGGCAGUCGGCCAAGGUGUUGGCCGAGGCGGCCAAAAAGGAUGUGCAGCGGAUCAGC .(((((.((..((((((((((((...((((((...((((((((((...((((....))))))))....)))---------------------)))..))).)))..))))))))))))..)).((.....)).))))).((......)) ( -55.10, z-score = -0.57, R) >droEre2.scaffold_4770 4590936 149 - 17746568 GGCGCAGUGAGUUGGCCAGCACCUCGGGCGACACGCCCCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((......(((((((((((((.((.((((..(((((...((((((.(...(((((((.((....)).)))))))...).))))))...))))).)))).)).))))))))))))).(((.((.....)).)))...)))....... ( -85.60, z-score = -4.75, R) >droYak2.chr3R 12783120 149 + 28832112 GGCGCAGUGAGUUGGCCAGCACCUCGGGCGACACGCCCCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((......(((((((((((((.((.((((..(((((...((((((.(...(((((((.((....)).)))))))...).))))))...))))).)))).)).))))))))))))).(((.((.....)).)))...)))....... ( -85.60, z-score = -4.75, R) >droSec1.super_0 7631676 149 - 21120651 GGCGCAGUGAGUUGGCCAGCACCUCGGGCGACACGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGCAAUGGGUCAGGGUCAGGGGCAGUCAGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((......(((((((((((((..((((((..(((.(...((((((.(.....(((((.((....)).))))).....).))))))...).))).))).))).))))))))))))).(((.((.....)).)))...)))....... ( -74.30, z-score = -2.29, R) >droSim1.chr3R 14590330 149 - 27517382 GGCGCAGUGAGUUGGCCAGCACCUCGGGUGACACGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACCAGCAGGGAAUGGGUCAGGGUCAGGGGCAGUCAGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((......(((((((((((((.((.((((..(((.(...((((((.(...(((((((.((....)).)))))))...).))))))...).))).)))).)).))))))))))))).(((.((.....)).)))...)))....... ( -79.30, z-score = -3.75, R) >droWil1.scaffold_181130 2402678 125 + 16660200 GGCGUAGUGAGUUGGCCAGCACCGCGGGCGACAACGGACGUUCGCUGAGCAGCUUUCUGCACGA-GUCG-----------------------AGCGAGUCUGCCAAGGUUCUGGCCGAGGCGGCUAAAAAUGACGUGGAGCGGAUAAGC .((.(((((..((((((((.(((...((((((......(((((((((.((((....)))).).)-).))-----------------------)))).))).)))..))).))))))))..).)))).......((.....)).....)) ( -44.30, z-score = 0.91, R) >droPer1.super_19 397753 113 - 1869541 GGCGUAGUGAGUUGGCCAGCACCUCGGGGGCCAA------------GAGCAGCUUUCUGCAGGAGUCCA------------------------GCGAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGAUGUGCAGCGGAUCAGC (((....((..(((((((((((((....((((..------------..((((....))))..((.((..------------------------..)).)))))).)))))))))))))..)))))......(((.((...)).)))... ( -48.10, z-score = -0.88, R) >dp4.chr2 25050732 113 - 30794189 GGCGUAGUGAGUUGGCCAGCACCUCGGGGGCCAA------------GAGCAGCUUUCUGCAGGAGUCCA------------------------GCGAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGAUGUGCAGCGGAUCAGC (((....((..(((((((((((((....((((..------------..((((....))))..((.((..------------------------..)).)))))).)))))))))))))..)))))......(((.((...)).)))... ( -48.10, z-score = -0.88, R) >droVir3.scaffold_13047 11132014 125 - 19223366 GACGUAGUGAGCUGGCCAGCACCUCGGGCGAUAAGCCGAGAUCGCUCAGCAGCUUUCUGCAGGA-GACA-----------------------AGCGAAUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAACGAUGUGCAGCGCAUUAGC ....(((((.((((((((((((((.((.((((..((.(((....))).((((....)))).(..-..).-----------------------.))..)))).)).)))))))))))......((((.......)).))))).))))).. ( -54.40, z-score = -2.12, R) >droMoj3.scaffold_6540 28098995 125 + 34148556 GACGUAGUGAGCUGGCCAGCACCUCAGGCGACAAGCCGCGAUCGCUGAGCAGCUUCCUGCAGGA-GUCA-----------------------AGCGAUUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAUGACGUGCAGCGCAUCAGC ......(((.((((((((((((((..(....)..((((.(((((((((((((....))))....-.)).-----------------------)))))))))))..)))))))))))...(((..((....)).)))..))).))).... ( -50.50, z-score = -0.10, R) >droGri2.scaffold_15074 1531349 125 + 7742996 GACGUAGUGAGCUGGCCAGCACCACGGGCGAUAAGCCGCCAUCGCUGAGCAGUUUCCUGCAGGA-GACG-----------------------GGCGAAUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAACGAUGUGCAGCGCAUUAGC ....(((((.(((((((((((((..((.((((..(((((((....)).)).((((((....)))-))).-----------------------)))..)))).))..))))))))))......((((.......)).))))).))))).. ( -57.40, z-score = -2.42, R) >consensus GGCGCAGUGAGUUGGCCAGCACCUCGGGCGACAAGCCGCGCUCCCUGAGCAGCUUCCUGCAGGACAACA_______________________GGCCAGUCGGCCAAGGUGCUGGCCGAGGCGGCCAAGAAGGACGUGCAGCGGAUCAGC ..(((.((....(((((((((((...(((...................((((....)))).(.....).................................)))..)))))))))))..(((.(.......).))))).)))....... (-34.97 = -34.68 + -0.29)
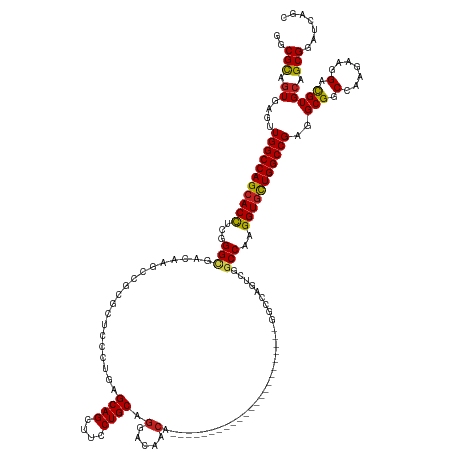
| Location | 8,495,923 – 8,496,072 |
|---|---|
| Length | 149 |
| Sequences | 12 |
| Columns | 149 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.44401 |
| G+C content | 0.65607 |
| Mean single sequence MFE | -59.69 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
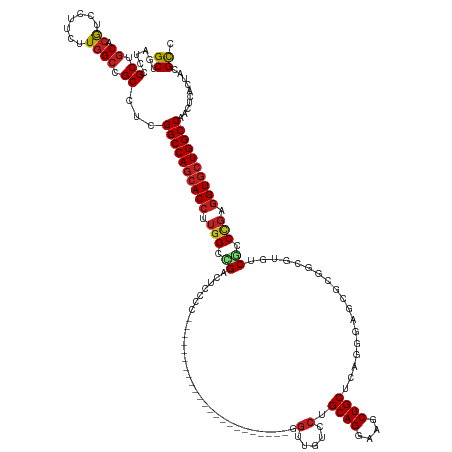
>dm3.chr3R 8495923 149 - 27905053 GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGUCUGCCCCUGACCCUGACCCAUUCCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGCGGCGUGUCGCCCGAGGUGCUGGCCAACUCACUGCGCC ........((.(((((.((.....)).))....((((((((((((((.((.((((((((..((((((.(..((.(((((.((....)).))))).))..).)))))))).).)))).).)).)))))))))))))).......))))). ( -68.40, z-score = -2.40, R) >droAna3.scaffold_13340 17017499 128 + 23697760 GCUGAUCCGCUGCACAUCCUUUUUGGCCGCCUCGGCCAACACCUUGGCCGACUGCCC---------------------CGUGUUGUCCUGCAGGAAGCUGCUCAAGGAACGGGGUGGAUCGCCGCCAGUGCUGGCCAACUCACUGCGCC ((.(((((((.((.((.......)))).)).((((((((....))))))))..((((---------------------(((....((((((((....))))...)))))))))))))))))).(((((((..(.....).))))).)). ( -55.10, z-score = -1.90, R) >droEre2.scaffold_4770 4590936 149 + 17746568 GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUGCCCCUGACCCUGACCCAUUCCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGGGGCGUGUCGCCCGAGGUGCUGGCCAACUCACUGCGCC ........((.(((((.((.....)).))....((((((((((((((.((((((((((...((((((.(..((.(((((.((....)).))))).))..).))))))...)))))).)))).)))))))))))))).......))))). ( -79.10, z-score = -4.72, R) >droYak2.chr3R 12783120 149 - 28832112 GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUGCCCCUGACCCUGACCCAUUCCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGGGGCGUGUCGCCCGAGGUGCUGGCCAACUCACUGCGCC ........((.(((((.((.....)).))....((((((((((((((.((((((((((...((((((.(..((.(((((.((....)).))))).))..).))))))...)))))).)))).)))))))))))))).......))))). ( -79.10, z-score = -4.72, R) >droSec1.super_0 7631676 149 + 21120651 GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCUGACUGCCCCUGACCCUGACCCAUUGCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGCGGCGUGUCGCCCGAGGUGCUGGCCAACUCACUGCGCC ........((.(((((.((.....)).))....((((((((((((((.(((((((((((..((((((.......(((((.((....)).))))).......)))))))).).)))).)))).)))))))))))))).......))))). ( -71.54, z-score = -3.01, R) >droSim1.chr3R 14590330 149 + 27517382 GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCUGACUGCCCCUGACCCUGACCCAUUCCCUGCUGGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGCGGCGUGUCACCCGAGGUGCUGGCCAACUCACUGCGCC ........((.(((((.((.....)).))....((((((((((((((.(((((((((((..((((((.(..((.(((((.((....)).))))).))..).)))))))).).)))).)))).)))))))))))))).......))))). ( -71.30, z-score = -3.64, R) >droWil1.scaffold_181130 2402678 125 - 16660200 GCUUAUCCGCUCCACGUCAUUUUUAGCCGCCUCGGCCAGAACCUUGGCAGACUCGCU-----------------------CGAC-UCGUGCAGAAAGCUGCUCAGCGAACGUCCGUUGUCGCCCGCGGUGCUGGCCAACUCACUACGCC ((......(((.............)))......((((((.(((..(((.(((.((..-----------------------((..-((((((((....))))...)))).))..))..))))))...))).))))))..........)). ( -36.42, z-score = 0.51, R) >droPer1.super_19 397753 113 + 1869541 GCUGAUCCGCUGCACAUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUCGC------------------------UGGACUCCUGCAGAAAGCUGCUC------------UUGGCCCCCGAGGUGCUGGCCAACUCACUACGCC ((......((.((.((.......)))).))...((((((((((((((((((.((..------------------------..)).))..((((....))))..------------..))))...))))))))))))..........)). ( -44.10, z-score = -2.29, R) >dp4.chr2 25050732 113 + 30794189 GCUGAUCCGCUGCACAUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUCGC------------------------UGGACUCCUGCAGAAAGCUGCUC------------UUGGCCCCCGAGGUGCUGGCCAACUCACUACGCC ((......((.((.((.......)))).))...((((((((((((((((((.((..------------------------..)).))..((((....))))..------------..))))...))))))))))))..........)). ( -44.10, z-score = -2.29, R) >droVir3.scaffold_13047 11132014 125 + 19223366 GCUAAUGCGCUGCACAUCGUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAUUCGCU-----------------------UGUC-UCCUGCAGAAAGCUGCUGAGCGAUCUCGGCUUAUCGCCCGAGGUGCUGGCCAGCUCACUACGUC (((.....((.((.((.......)))).))...((((((((((((((.((((..(((-----------------------((.(-((..((((....)))).))))).....)))..)))).))))))))))))))))).......... ( -54.50, z-score = -3.08, R) >droMoj3.scaffold_6540 28098995 125 - 34148556 GCUGAUGCGCUGCACGUCAUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAAUCGCU-----------------------UGAC-UCCUGCAGGAAGCUGCUCAGCGAUCGCGGCUUGUCGCCUGAGGUGCUGGCCAGCUCACUACGUC (((((.(((......((((....))))))).))(((((((((((..(..((.(((((-----------------------.((.-....((((....)))))))))))))((((....)))))..)))))))))))))).......... ( -54.90, z-score = -1.70, R) >droGri2.scaffold_15074 1531349 125 - 7742996 GCUAAUGCGCUGCACAUCGUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGAUUCGCC-----------------------CGUC-UCCUGCAGGAAACUGCUCAGCGAUGGCGGCUUAUCGCCCGUGGUGCUGGCCAGCUCACUACGUC (((.....((.((.((.......)))).))...((((((((((.(((.((((..(((-----------------------.(((-((((((((....))))..)).)).))))))..)))).))).))))))))))))).......... ( -57.70, z-score = -3.48, R) >consensus GCUGAUCCGCUGCACGUCCUUCUUGGCCGCCUCGGCCAGCACCUUGGCCGACUCCCC_______________________GGUUGUCCUGCAGGAAGCUGCUCAGGGAGCGCGGCGUGUCGCCCGAGGUGCUGGCCAACUCACUACGCC ((......((.((.((.......)))).))...((((((((((.(((.((...............................(....)..((((....))))..................)).))).))))))))))..........)). (-32.70 = -32.88 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:47 2011