| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,435,578 – 8,435,670 |
| Length | 92 |
| Max. P | 0.896318 |

| Location | 8,435,578 – 8,435,670 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.57648 |
| G+C content | 0.56639 |
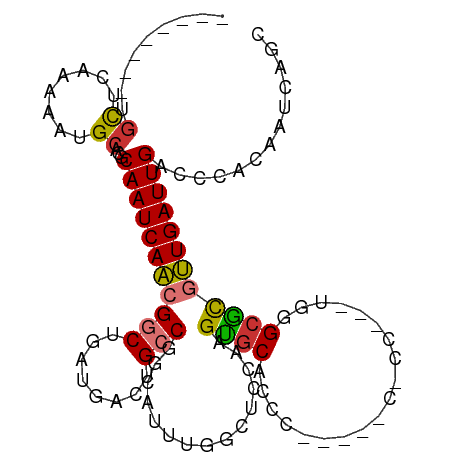
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.51 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
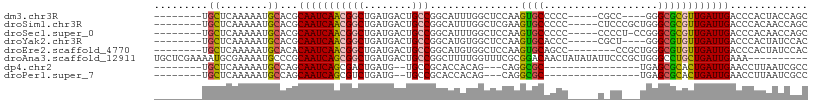
>dm3.chr3R 8435578 92 + 27905053 --------UGCUCAAAAAUGCACGCAAUCAACGGCUGAUGACUGCCGGCAUUUGGCUCCAAGUGCCCCC-----CGCC----GGGCGCGUUGAUUGACCCACUACCAGC --------(((........)))..(((((((((((........)))((((((((....))))))))(((-----....----)))...))))))))............. ( -31.80, z-score = -1.59, R) >droSim1.chr3R 14526891 96 - 27517382 --------UGCUCAAAAAUGCACGCAAUCAACGGCUGAUGACUGCCGGCAUUUGGCUCGAAGUGCCCCC-----CUCCCGCUGGGCGCGUUGAUUGACCCACAACCAGC --------(((........)))..(((((((((((........)))(((.....)))....((((((..-----(....)..))))))))))))))............. ( -28.60, z-score = -0.33, R) >droSec1.super_0 7568902 95 - 21120651 --------UGCUCAAAAAUGCACGCAAUCAACGGCUGAUGACUGCCGGCAUUUGGCUCCAAGUGCCCCC-----CCCCU-CCGGGCGCGUUGAUUGACCCACAACCAGC --------(((........)))..(((((((((((........)))((((((((....)))))))).((-----(....-..)))...))))))))............. ( -32.00, z-score = -2.09, R) >droYak2.chr3R 12721640 92 + 28832112 --------UGCUCAAAAAUGCACGCAAUCAACGGCUGAUGACUGCCGGCAUGUGGCUCCAAGUGCACCC-----CGCU----GGGCGUGUUGAUUGACCCACUAUCCAC --------(((........)))..(((((((((((........))).((((.((....)).))))((((-----(...----))).))))))))))............. ( -26.10, z-score = 0.08, R) >droEre2.scaffold_4770 4529945 93 - 17746568 --------UGCUCAAAAAUGCACACAAUCAACGGCUGAUGACUGCCGGCAUGUGGCUCCAAGUGCAGCC--------CCGCUGGGCGUGUUGAUUGACCCACUAUCCAC --------(((........)))..(((((((((((........))).((((.((....)).)))).(((--------(....))))..))))))))............. ( -30.40, z-score = -1.14, R) >droAna3.scaffold_12911 3368269 99 + 5364042 UGCUCGAAAAUGCGAAAAUGCCCGCAAUCAGCGGCUGAUGACUGCCGGCUUUUGGUUUCGCGGACAACUAUAUAUUCCCGCUGGGCCUGCUGAUUGAAA---------- .((........))...........(((((((((((........))).......((((..((((..............))))..)))).))))))))...---------- ( -30.04, z-score = -1.04, R) >dp4.chr2 2541842 80 + 30794189 --------UGCUCAAAAAUGCCAGCAAUCAGCGACUGAUG--UGCCGCACCACAG---CAGGCGC----------------UGAGCGCACUGAUUGAACCUUAAUCGCC --------.((............((..((((((.(((.((--((......)))).---))).)))----------------)))..))...((((((...)))))))). ( -24.30, z-score = -1.25, R) >droPer1.super_7 2712624 80 + 4445127 --------UGCUCAAAAAUGCCAGCAAUCAGCGUCUGAUG--UGCCGCACCACAG---CAGGCGC----------------UGAGCGCACUGAUUGAACCUUAAUCGCC --------.((............((..((((((((((.((--((......)))).---)))))))----------------)))..))...((((((...)))))))). ( -28.70, z-score = -2.49, R) >consensus ________UGCUCAAAAAUGCACGCAAUCAACGGCUGAUGACUGCCGGCAUUUGGCUCCAAGUGCACCC_____C_CC___UGGGCGCGUUGAUUGACCCACAAUCAGC .........((........))...(((((((((((........)))...............((((...................))))))))))))............. (-15.95 = -15.51 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:37 2011