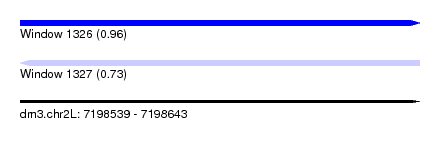
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,198,539 – 7,198,643 |
| Length | 104 |
| Max. P | 0.961311 |

| Location | 7,198,539 – 7,198,643 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 59.46 |
| Shannon entropy | 0.77723 |
| G+C content | 0.36224 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -6.54 |
| Energy contribution | -6.29 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
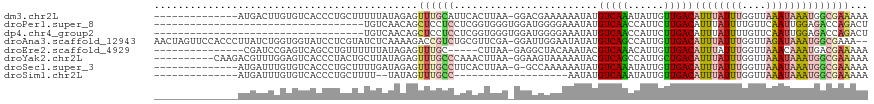
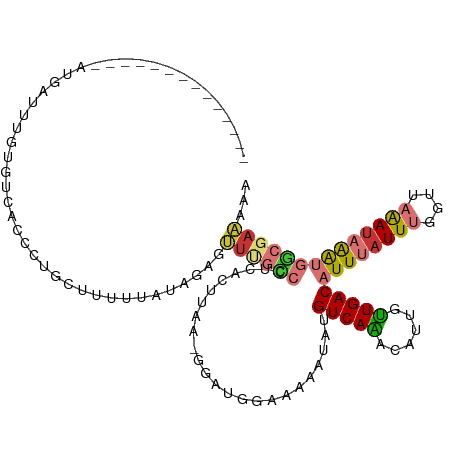
>dm3.chr2L 7198539 104 + 23011544 UUUUUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUUUUUUCGUCC-UUAAGUGAAUGCAAACUCUAUAAAAAGCAGGGUGACACAAGUCAU-------------- ......((.(((((((((((.(((...(((((((......)))))))....))).))..-.)))))))))))..(((((.........)))))(((....)))..-------------- ( -17.60, z-score = -1.72, R) >droPer1.super_8 905382 84 - 3966273 AGUCUGGUCUCCAAUUGAACAAAAUAAAUGUCAAGAAUGGUUGACAUAUUUCCCCAUCCACCCACCGAGGAGGAGCUGUUGACA----------------------------------- .(((.((.((((...............(((((((......))))))).........(((.........)))))))))...))).----------------------------------- ( -18.10, z-score = 0.04, R) >dp4.chr4_group2 840833 84 + 1235136 AGUCUGGUCUCCAAUUGAACAAAAUAAAUGUCAAGAAUGGUUGACAUAUUUCCCCAUCCACCCACCGAGGAGGAGCUGUUGACA----------------------------------- .(((.((.((((...............(((((((......))))))).........(((.........)))))))))...))).----------------------------------- ( -18.10, z-score = 0.04, R) >droAna3.scaffold_12943 1052024 116 - 5039921 --UUUCGCCAUUUAUCUAACCAAAUAAAUGUCAACAAUGGCUGACAUAUAUUCCAAUCC-UCGAACGCAGACGGUCUUUUGAGAUACGAGGAUACCACCAGAUAAGGGUGGAACUAGUU --(((((((.(((((((.....((((.((((((........)))))).))))...((((-(((..((....))((((....)))).)))))))......))))))))))))))...... ( -32.10, z-score = -3.13, R) >droEre2.scaffold_4929 16117888 98 + 26641161 UUUUUCGUCAUUUGUUUAACCAAAUAAAUGUCAACAAUGUUUGACGUAUUUGUAGCCUC-UUAAG-----GCAAACUCUAUAAAAAACAGGCUGACUCGGAUCG--------------- ......(((((((((((...((((((...(((((......))))).))))))..((((.-...))-----))............))))))).))))........--------------- ( -21.50, z-score = -2.60, R) >droYak2.chr2L 16620161 108 - 22324452 UUUUUCGCCAUUUAUUUAACCAAAUAAAUGUCAGCAAUGGCUGACGUAUUUUUACUUCC-UUAAGUUUGGGCAAACUCUAUAAGCAGUAGGGUGACUCCAAACGUCUUG---------- .........((((((((....))))))))((((((....))))))(((....)))....-....(((((((...(((((((.....)))))))...)))))))......---------- ( -28.50, z-score = -3.22, R) >droSec1.super_3 2722128 103 + 7220098 UUUUUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUUUUUUGGC-C-UUAAGUGAAGGCAAACUCUAUCAAAAGCAGGGUGACACAAAUCAU-------------- ......(((.((((((((((((((...(((((((......)))))))....))))).-.-))))))))))))..(((((.........)))))............-------------- ( -23.70, z-score = -3.43, R) >droSim1.chr2L 6988451 84 + 22036055 UUUUUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUAUUUGACAUAUU-------------------GGCAAACUAUA--AAAAGCAGGGUGACACAAAUCAU-------------- ......((((.((((((....))))))(((((((......)))))))..)-------------------)))........--........(....).........-------------- ( -14.40, z-score = -2.23, R) >consensus UUUUUCGCCAUUUAUUUAACCAAAUAAAUGUCAACAAUGGUUGACAUAUUUUUCCAUCC_UUAAGCGAAGGCAAACUCUAUAAAAAGCAGGGUGACACAAAUCAU______________ ...........................(((((((......)))))))........................................................................ ( -6.54 = -6.29 + -0.25)

| Location | 7,198,539 – 7,198,643 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 59.46 |
| Shannon entropy | 0.77723 |
| G+C content | 0.36224 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -8.36 |
| Energy contribution | -8.85 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7198539 104 - 23011544 --------------AUGACUUGUGUCACCCUGCUUUUUAUAGAGUUUGCAUUCACUUAA-GGACGAAAAAAUAUGUCAAAUAUUGUUGACAUUUAUUUGGUUAAAUAAAUGGCGAAAAA --------------.((((....))))...((((.(((((.((((....))))......-.(((.(((....(((((((......)))))))...))).)))..))))).))))..... ( -19.10, z-score = -0.80, R) >droPer1.super_8 905382 84 + 3966273 -----------------------------------UGUCAACAGCUCCUCCUCGGUGGGUGGAUGGGGAAAUAUGUCAACCAUUCUUGACAUUUAUUUUGUUCAAUUGGAGACCAGACU -----------------------------------.(((.....(((((((.....)))(((((..((....(((((((......)))))))...))..)))))...))))....))). ( -20.90, z-score = 0.22, R) >dp4.chr4_group2 840833 84 - 1235136 -----------------------------------UGUCAACAGCUCCUCCUCGGUGGGUGGAUGGGGAAAUAUGUCAACCAUUCUUGACAUUUAUUUUGUUCAAUUGGAGACCAGACU -----------------------------------.(((.....(((((((.....)))(((((..((....(((((((......)))))))...))..)))))...))))....))). ( -20.90, z-score = 0.22, R) >droAna3.scaffold_12943 1052024 116 + 5039921 AACUAGUUCCACCCUUAUCUGGUGGUAUCCUCGUAUCUCAAAAGACCGUCUGCGUUCGA-GGAUUGGAAUAUAUGUCAGCCAUUGUUGACAUUUAUUUGGUUAGAUAAAUGGCGAAA-- ........(((((.......))))).(((((((..(((....))).((....))..)))-))))..(((((.((((((((....)))))))).))))).((((......))))....-- ( -28.70, z-score = -0.95, R) >droEre2.scaffold_4929 16117888 98 - 26641161 ---------------CGAUCCGAGUCAGCCUGUUUUUUAUAGAGUUUGC-----CUUAA-GAGGCUACAAAUACGUCAAACAUUGUUGACAUUUAUUUGGUUAAACAAAUGACGAAAAA ---------------.....((.((((((.(((((............((-----((...-.))))............)))))..))))))..(((((((......)))))))))..... ( -20.35, z-score = -1.40, R) >droYak2.chr2L 16620161 108 + 22324452 ----------CAAGACGUUUGGAGUCACCCUACUGCUUAUAGAGUUUGCCCAAACUUAA-GGAAGUAAAAAUACGUCAGCCAUUGCUGACAUUUAUUUGGUUAAAUAAAUGGCGAAAAA ----------...(((((..((......))...(((((...((((((....))))))..-..))))).....))))).((((((..((((.........))))....))))))...... ( -22.70, z-score = -0.92, R) >droSec1.super_3 2722128 103 - 7220098 --------------AUGAUUUGUGUCACCCUGCUUUUGAUAGAGUUUGCCUUCACUUAA-G-GCCAAAAAAUAUGUCAAAUAUUGUUGACAUUUAUUUGGUUAAAUAAAUGGCGAAAAA --------------.((((....))))....(((((....)))))(((((.........-(-((((((....(((((((......)))))))...)))))))........))))).... ( -21.73, z-score = -1.57, R) >droSim1.chr2L 6988451 84 - 22036055 --------------AUGAUUUGUGUCACCCUGCUUUU--UAUAGUUUGCC-------------------AAUAUGUCAAAUAUUGUUGACAUUUAUUUGGUUAAAUAAAUGGCGAAAAA --------------.((((....))))...((((.((--(((.....(((-------------------((.(((((((......)))))))....)))))...))))).))))..... ( -18.40, z-score = -2.20, R) >consensus ______________AUGAUUUGUGUCACCCUGCUUUUUAUAGAGUUUGCCCUCACUUAA_GGAUGGAAAAAUAUGUCAAACAUUGUUGACAUUUAUUUGGUUAAAUAAAUGGCGAAAAA ............................................((((((........................(((((......)))))((((((((....))))))))))))))... ( -8.36 = -8.85 + 0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:10 2011