
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,421,540 – 8,421,718 |
| Length | 178 |
| Max. P | 0.849205 |

| Location | 8,421,540 – 8,421,650 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.84 |
| Shannon entropy | 0.08019 |
| G+C content | 0.56941 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -26.99 |
| Energy contribution | -26.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
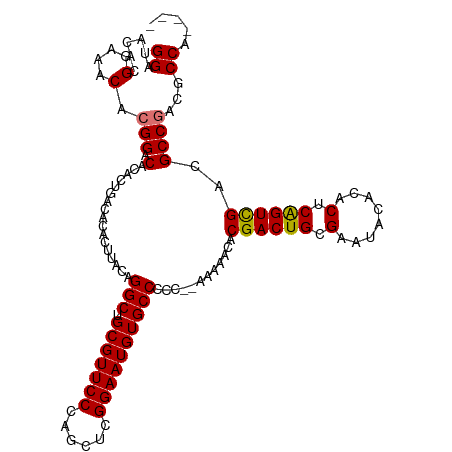
>dm3.chr3R 8421540 110 + 27905053 GGAUACACGGAAACACGGACACAAUGACACACUUACAGGCUGCGUUCCCAGCUCGGAAUGUGCCCCC--AAAAACACGACUGCGAAUACACACUCGGUCGACGCCGACGCCA---- ((......(....).(((.(.................(((.(((((((......))))))))))...--.......((((((.(........).))))))..))))...)).---- ( -28.10, z-score = -1.02, R) >droSim1.chr3R 14512743 112 - 27517382 GGAUACACGGAAACACGGACACACUGACACACUUACAGGCUGCGUUCCCAGCUCGGAAUGUGCCCCCGAAAAAACACGACUGCGAAUACACACUCAGUCGACGCCAACGCCA---- ((.....(((.....(((.....)))...........(((.(((((((......)))))))))).)))........((((((.(........).))))))...)).......---- ( -27.40, z-score = -1.34, R) >droYak2.chr3R 12707439 110 + 28832112 GGAUACACGGAAACACGGACACACUGACACACUUACAGGCUGCGUUCCCAGCUCGGAAUGUGCCCCC--AAAAACACGACUGCGAAUACACACUCAGUCGACGCCGACGCCG---- ((......(....).(((.(.................(((.(((((((......))))))))))...--.......((((((.(........).))))))..))))...)).---- ( -28.80, z-score = -1.69, R) >droEre2.scaffold_4770 4515812 114 - 17746568 GGAUACACGGAAACACGGACACACUGACACACUUACAGGCUGCGUUCCCAGCUCGGAAUGUGCCCCC--AAAAACACGACUGCGAAUACACACUCGGUUGACGCCGACGCCGCCGC ((......(....).(((.(.................(((.(((((((......))))))))))...--.......((((((.(........).))))))..))))...))..... ( -26.00, z-score = 0.51, R) >consensus GGAUACACGGAAACACGGACACACUGACACACUUACAGGCUGCGUUCCCAGCUCGGAAUGUGCCCCC__AAAAACACGACUGCGAAUACACACUCAGUCGACGCCGACGCCA____ ((......(....).(((.(.................(((.(((((((......))))))))))............((((((.(........).))))))..))))...))..... (-26.99 = -26.80 + -0.19)

| Location | 8,421,614 – 8,421,711 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Shannon entropy | 0.20005 |
| G+C content | 0.64252 |
| Mean single sequence MFE | -49.24 |
| Consensus MFE | -40.52 |
| Energy contribution | -40.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8421614 97 - 27905053 GCAACAUGCGCGCAGCCGACGUCGCUGUUGAAGCUCUGCAGCAGCUGCCGAGCAGAGGCGUU------------------GGCGUCGGCGUCGACCGAGUGUGUAUUCGCAGUCG ((...((((((((.((((((((((.((((...((((.((((...)))).))))...)))).)------------------))))))))).((....))))))))))..))..... ( -45.00, z-score = -1.02, R) >droSim1.chr3R 14512819 96 + 27517382 GCAACAUGCGCGCAGCCGACGUCGCUGUUGUAGCUCUGCAGCAGCUG-CGAGCAGAGGCGUU------------------GGCGUUGGCGUCGACUGAGUGUGUAUUCGCAGUCG ((...((((((((.(((((((((.((((((((((((....).)))))-).))))).))))))------------------)))((((....))))...))))))))..))..... ( -45.20, z-score = -1.11, R) >droSec1.super_0 7555181 96 + 21120651 GCAACAUGCGCGCAGUCGACGUCGCUGUUGUAGCUUUGCAGCAGCUG-CGAGCAGAGGCGUU------------------GGCGUCGGCGUCGACUGAGUGUGUAUUCGCAGUCG ((((.((((((.((((((((((((((((((((....))))))))).(-(.(((......)))------------------.))...))))))))))).)))))).)).))..... ( -47.10, z-score = -2.10, R) >droYak2.chr3R 12707513 102 - 28832112 GCAACAUGCGCGCAGCCGACGUCGCUGUCGAAGCUCUGCAGCAGCUG-CGGGCAGAGGCGUUGGCUU------------CGGCGUCGGCGUCGACUGAGUGUGUAUUCGCAGUCG ((((.((((((.(((.((((((((.((((((((((..((.((..(((-....)))..)))).)))))------------))))).)))))))).))).)))))).)).))..... ( -50.80, z-score = -1.57, R) >droEre2.scaffold_4770 4515886 114 + 17746568 GCAACAUGCGCGCAGCCGACGUCGCUGUCGCAGCUCUGCAGCAGCAG-CGAGCAGAGGCGUUGGCUUUGGCGUUGGCGGCGGCGUCGGCGUCAACCGAGUGUGUAUUCGCAGUCG ((...((((((((.(((((((((((((((....((((((.((....)-)..))))))(((((......))))).))))))))))))))).((....))))))))))..))..... ( -58.10, z-score = -1.74, R) >consensus GCAACAUGCGCGCAGCCGACGUCGCUGUUGAAGCUCUGCAGCAGCUG_CGAGCAGAGGCGUU__________________GGCGUCGGCGUCGACUGAGUGUGUAUUCGCAGUCG ((...((((((((.((((((((((((((((((....)))))))))......((....)).....................))))))))).((....))))))))))..))..... (-40.52 = -40.92 + 0.40)


| Location | 8,421,621 – 8,421,718 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.29 |
| Shannon entropy | 0.20701 |
| G+C content | 0.61269 |
| Mean single sequence MFE | -48.58 |
| Consensus MFE | -38.38 |
| Energy contribution | -38.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8421621 97 - 27905053 UUUGAGUGCAACAUGCGCGCAGCCGACGUCGCUGUUGAAGCUCUGCAGCAGCUGCCGAGCAGAGGCGUU------------------GGCGUCGGCGUCGACCGAGUGUGUAUUC ...(((((((.(((.((((..((((((((((.((((...((((.((((...)))).))))...)))).)------------------)))))))))..))..)).)))))))))) ( -43.60, z-score = -1.32, R) >droSim1.chr3R 14512826 96 + 27517382 UUUGAGUGCAACAUGCGCGCAGCCGACGUCGCUGUUGUAGCUCUGCAGCAGCUG-CGAGCAGAGGCGUU------------------GGCGUUGGCGUCGACUGAGUGUGUAUUC ...(((((((.(((.(((((.(((((((((.((((((((((((....).)))))-).))))).))))))------------------)))....))).)).....)))))))))) ( -45.60, z-score = -2.09, R) >droSec1.super_0 7555188 96 + 21120651 UUUGAGUGCAACAUGCGCGCAGUCGACGUCGCUGUUGUAGCUUUGCAGCAGCUG-CGAGCAGAGGCGUU------------------GGCGUCGGCGUCGACUGAGUGUGUAUUC ...((((((.....((((.((((((((((((((((((((....))))))))).(-(.(((......)))------------------.))...))))))))))).)))))))))) ( -46.80, z-score = -2.83, R) >droYak2.chr3R 12707520 102 - 28832112 UUUGAGUGCAACAUGCGCGCAGCCGACGUCGCUGUCGAAGCUCUGCAGCAGCUG-CGGGCAGAGGCGUU------------GGCUUCGGCGUCGGCGUCGACUGAGUGUGUAUUC ...((((((.....((((.(((.((((((((.((((((((((..((.((..(((-....)))..)))).------------)))))))))).)))))))).))).)))))))))) ( -50.50, z-score = -2.13, R) >droEre2.scaffold_4770 4515893 114 + 17746568 UUUGAGUGCAACAUGCGCGCAGCCGACGUCGCUGUCGCAGCUCUGCAGCAGCAG-CGAGCAGAGGCGUUGGCUUUGGCGUUGGCGGCGGCGUCGGCGUCAACCGAGUGUGUAUUC ...(((((((.(((.((....(((((((((((((((....((((((.((....)-)..))))))(((((......))))).)))))))))))))))......)).)))))))))) ( -56.40, z-score = -1.68, R) >consensus UUUGAGUGCAACAUGCGCGCAGCCGACGUCGCUGUUGAAGCUCUGCAGCAGCUG_CGAGCAGAGGCGUU__________________GGCGUCGGCGUCGACUGAGUGUGUAUUC ...(((((((.(((...((..((((((((((((((((((....)))))))))......((....)).....................)))))))))..)).....)))))))))) (-38.38 = -38.98 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:35 2011