
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,377,236 – 8,377,335 |
| Length | 99 |
| Max. P | 0.997760 |

| Location | 8,377,236 – 8,377,335 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.40 |
| Shannon entropy | 0.20473 |
| G+C content | 0.48201 |
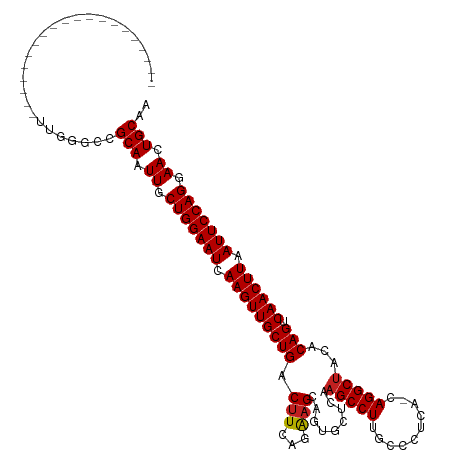
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -29.21 |
| Energy contribution | -29.19 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
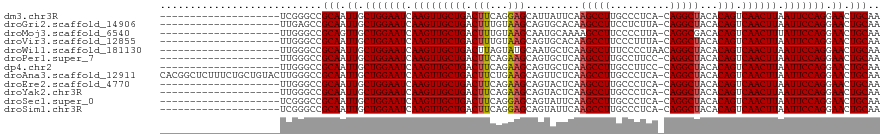
>dm3.chr3R 8377236 99 + 27905053 --------------------UCGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGGAGCAUUAUUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((......(((......))).(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -32.80, z-score = -2.67, R) >droGri2.scaffold_14906 12472020 99 - 14172833 --------------------UUGAGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUUGUAAGCAGUGCACAAGCCUUCCUCUUA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((....((((.....))))..(((((........-.)))))...))).)))))).))))))).)).))).. ( -32.50, z-score = -2.62, R) >droMoj3.scaffold_6540 12381815 99 + 34148556 --------------------UUGGGCCGCAGUUGCUGGAAUCAAGUUGCUGACUUUGUAAGCAAUGCAAAAGCCUUCCCCUUA-CAGGCGACACAGUCAACUUUAUUCCAGGAACUGCAA --------------------.......((((((.(((((((.(((((((((..((((((.....)))))).((((........-.))))....))).)))))).))))))).)))))).. ( -39.80, z-score = -4.37, R) >droVir3.scaffold_12855 9576631 99 - 10161210 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUUGUAAGCAGUGCACAAGCCUUCCCUUUA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((....((((.....))))..(((((........-.)))))...))).)))))).))))))).)).))).. ( -32.50, z-score = -2.20, R) >droWil1.scaffold_181130 2186214 100 + 16660200 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUAGUAUGCAAUGCUCAAGCCUUUCCCCUAACAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((....(((((...)))))..(((((..........)))))...))).)))))).))))))).)).))).. ( -32.20, z-score = -2.61, R) >droPer1.super_7 2652975 99 + 4445127 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGAAGCAGUGCUCAAGCCUUGCCUUCC-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((.(((..(((((((.((....)).))).)))).-.))).....))).)))))).))))))).)).))).. ( -32.80, z-score = -1.58, R) >dp4.chr2 2484985 99 + 30794189 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGAAGCAGUGCUCAAGCCUUGCCUUCC-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((.(((..(((((((.((....)).))).)))).-.))).....))).)))))).))))))).)).))).. ( -32.80, z-score = -1.58, R) >droAna3.scaffold_12911 3307981 119 + 5364042 CACGGCUCUUUCUGCUGUACUUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCUGAAGCAGUUCUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA ...(((((..............)))))(((.((.(((((((.(((((((((.....(((........)))(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -39.64, z-score = -2.33, R) >droEre2.scaffold_4770 4470918 99 - 17746568 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGAAGCAGUACUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.((((((((((((.(....).))).....(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -32.10, z-score = -2.07, R) >droYak2.chr3R 12662242 99 + 28832112 --------------------UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGAAGCAGUACUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.((((((((((((.(....).))).....(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -32.10, z-score = -2.07, R) >droSec1.super_0 7511310 99 - 21120651 --------------------UCGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGGAGCAGUAUUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((......(((......))).(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -32.80, z-score = -2.21, R) >droSim1.chr3R 14466423 99 - 27517382 --------------------UCGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGGAGCAGUAUUCAAGCCUUGCCCUCA-CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA --------------------.......(((.((.(((((((.(((((((((......(((......))).(((((((....))-.)))))...))).)))))).))))))).)).))).. ( -32.80, z-score = -2.21, R) >consensus ____________________UUGGGCCGCAAUUGCUGGAAUCAAGUUGCUGACUUCAGAAGCAGUGCUCAAGCCUUGCCCUCA_CAGGCUACACAGUCAACUUAAUUCCAGGAACUGCAA ...........................(((.((.(((((((.(((((((((.(((...))).........(((((..........)))))...))).)))))).))))))).)).))).. (-29.21 = -29.19 + -0.02)
| Location | 8,377,236 – 8,377,335 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.40 |
| Shannon entropy | 0.20473 |
| G+C content | 0.48201 |
| Mean single sequence MFE | -40.09 |
| Consensus MFE | -36.07 |
| Energy contribution | -36.12 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
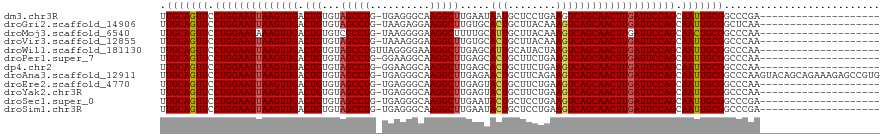
>dm3.chr3R 8377236 99 - 27905053 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAAUAAUGCUCCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCGA-------------------- .(((((((.((((((((((((((.(((....((((.-...))))..(((.........)))........))))))))))))))))).)))))))......-------------------- ( -38.40, z-score = -3.40, R) >droGri2.scaffold_14906 12472020 99 + 14172833 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UAAGAGGAAGGCUUGUGCACUGCUUACAAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCUCAA-------------------- .(((((((.((((((((((((((.(((.......((-((((....((((....)).)).))))))....))))))))))))))))).)))))))......-------------------- ( -39.50, z-score = -3.65, R) >droMoj3.scaffold_6540 12381815 99 - 34148556 UUGCAGUUCCUGGAAUAAAGUUGACUGUGUCGCCUG-UAAGGGGAAGGCUUUUGCAUUGCUUACAAAGUCAGCAACUUGAUUCCAGCAACUGCGGCCCAA-------------------- .(((((((.(((((((.((((((.(((...(...((-((((.((...((....)).)).))))))..).))))))))).))))))).)))))))......-------------------- ( -38.10, z-score = -3.10, R) >droVir3.scaffold_12855 9576631 99 + 10161210 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UAAAGGGAAGGCUUGUGCACUGCUUACAAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((..((..((((((.-........)))).))..))(((((.....)).))))))))))))))))).)))))))......-------------------- ( -38.60, z-score = -3.35, R) >droWil1.scaffold_181130 2186214 100 - 16660200 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUGUUAGGGGAAAGGCUUGAGCAUUGCAUACUAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((.(((.(((((.((((..((......))..))))..)).))).....))))))))))))))))).)))))))......-------------------- ( -37.60, z-score = -2.96, R) >droPer1.super_7 2652975 99 - 4445127 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-GGAAGGCAAGGCUUGAGCACUGCUUCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((.((.....((((.-...))))..(((((.((........)).))))))))))))))))))))).)))))))......-------------------- ( -42.00, z-score = -3.39, R) >dp4.chr2 2484985 99 - 30794189 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-GGAAGGCAAGGCUUGAGCACUGCUUCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((.((.....((((.-...))))..(((((.((........)).))))))))))))))))))))).)))))))......-------------------- ( -42.00, z-score = -3.39, R) >droAna3.scaffold_12911 3307981 119 - 5364042 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAGAACUGCUUCAGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAAGUACAGCAGAAAGAGCCGUG ..((((((.((((((((((((((.((.....((((.-...))))..(((((..(((....)))..))))))))))))))))))))).))))))(((.(..............).)))... ( -43.34, z-score = -2.43, R) >droEre2.scaffold_4770 4470918 99 + 17746568 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAGUACUGCUUCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((.((.....((((.-...))))..(((((.((........)).))))))))))))))))))))).)))))))......-------------------- ( -42.40, z-score = -4.04, R) >droYak2.chr3R 12662242 99 - 28832112 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAGUACUGCUUCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA-------------------- .(((((((.((((((((((((((.((.....((((.-...))))..(((((.((........)).))))))))))))))))))))).)))))))......-------------------- ( -42.40, z-score = -4.04, R) >droSec1.super_0 7511310 99 + 21120651 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAAUACUGCUCCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCGA-------------------- .(((((((.((((((((((((((.(((....((((.-...))))..(((.........)))........))))))))))))))))).)))))))......-------------------- ( -38.40, z-score = -3.12, R) >droSim1.chr3R 14466423 99 + 27517382 UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG-UGAGGGCAAGGCUUGAAUACUGCUCCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCGA-------------------- .(((((((.((((((((((((((.(((....((((.-...))))..(((.........)))........))))))))))))))))).)))))))......-------------------- ( -38.40, z-score = -3.12, R) >consensus UUGCAGUUCCUGGAAUUAAGUUGACUGUGUAGCCUG_UGAGGGCAAGGCUUGAGCACUGCUUCUGAAGUCAGCAACUUGAUUCCAGCAAUUGCGGCCCAA____________________ .(((((((.((((((((((((((.(((...(((((..........))))).....(((........)))))))))))))))))))).))))))).......................... (-36.07 = -36.12 + 0.05)
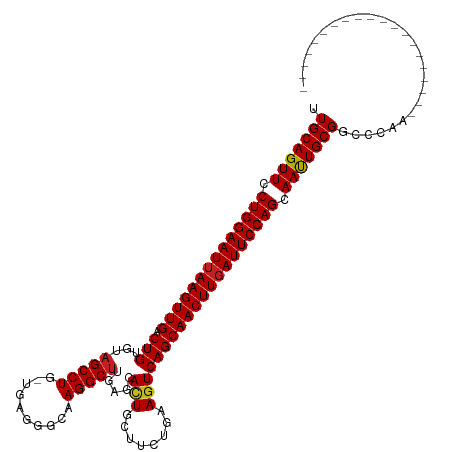
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:26 2011