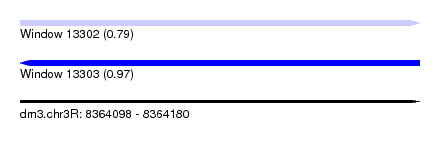
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,364,098 – 8,364,180 |
| Length | 82 |
| Max. P | 0.970879 |

| Location | 8,364,098 – 8,364,180 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Shannon entropy | 0.44118 |
| G+C content | 0.45508 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -13.21 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
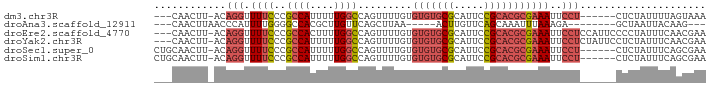
>dm3.chr3R 8364098 82 + 27905053 UUUACUAAAAUAGAG------AGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAAUGGCGGGAAAACCUGU-AAGUUG--- ........(((..(.------(((......(.(((((.....))))).)......((.((((....)))).))....))).)-..))).--- ( -20.40, z-score = -1.69, R) >droAna3.scaffold_12911 3295018 73 + 5364042 ---CUUGUAAUUAGC--------UCUUUAAAUUUGCUGAACAAGU-----UUAAGCUGAACAAGCGUGGCCCCAAAAUGGGUUAAGUUG--- ---(((((..(((((--------(...((((((((.....)))))-----))))))))))))))..((((((......)))))).....--- ( -19.70, z-score = -2.21, R) >droEre2.scaffold_4770 4458015 88 - 17746568 UUCGUUGAAAUAGGGGAAUGGAGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAGUGGCGGGAAAACCUGU-AAGUUG--- .........((((((((((......)))))(.(((((.....))))).)......((.((((....)))).))....)))))-......--- ( -24.10, z-score = -1.27, R) >droYak2.chr3R 12649166 88 + 28832112 UUCGUUGAAAUAGAGGAAUAGAGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAAUGGCGGGAAAACCUGU-AAGUUG--- (((.((......)).)))((.(((......(.(((((.....))))).)......((.((((....)))).))....))).)-).....--- ( -22.30, z-score = -1.40, R) >droSec1.super_0 7498610 85 - 21120651 UUCGCUGAAAUAGAG------AGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAAUGGCGGGAAAACCUGU-AAGUUGCAG ....(((.(((..(.------(((......(.(((((.....))))).)......((.((((....)))).))....))).)-..))).))) ( -23.20, z-score = -1.25, R) >droSim1.chr3R 14451679 85 - 27517382 UUCGCUGAAAUAGAG------AGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAAUGGCGGGAAAACCUGU-AAGUUGCAG ....(((.(((..(.------(((......(.(((((.....))))).)......((.((((....)))).))....))).)-..))).))) ( -23.20, z-score = -1.25, R) >consensus UUCGCUGAAAUAGAG______AGGAAUUUCGCGUGCGGAAUGCGCACACACAAAACUGGCCAAAAAUGGCGGGAAAACCUGU_AAGUUG___ ................................(((((.....)))))......((((.((((....))))(((....)))....)))).... (-13.21 = -12.93 + -0.27)
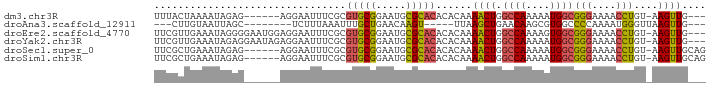
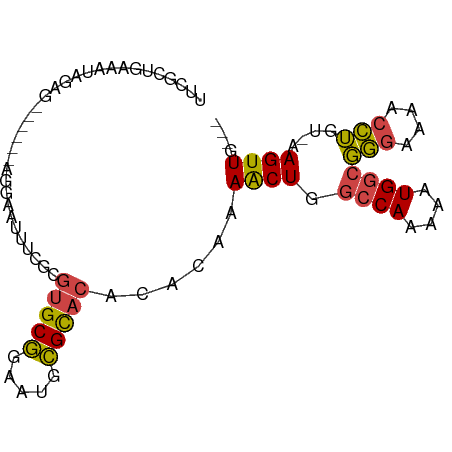
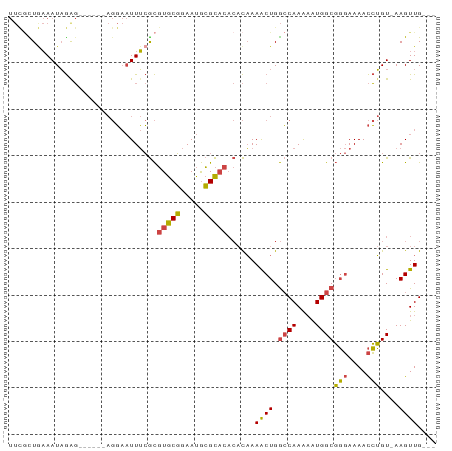
| Location | 8,364,098 – 8,364,180 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Shannon entropy | 0.44118 |
| G+C content | 0.45508 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -12.93 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
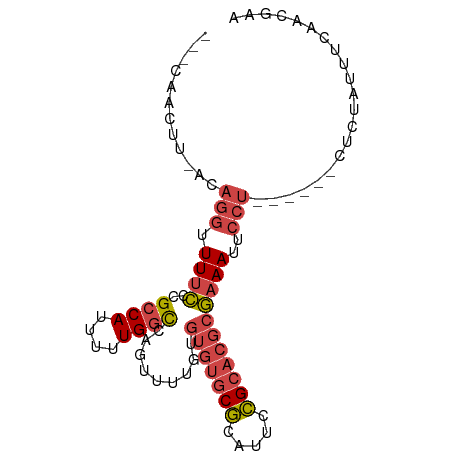
>dm3.chr3R 8364098 82 - 27905053 ---CAACUU-ACAGGUUUUCCCGCCAUUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCU------CUCUAUUUUAGUAAA ---....((-((.((.....))((((....)))).(((((((((((((.......)))))))))))))...------..........)))). ( -21.00, z-score = -2.87, R) >droAna3.scaffold_12911 3295018 73 - 5364042 ---CAACUUAACCCAUUUUGGGGCCACGCUUGUUCAGCUUAA-----ACUUGUUCAGCAAAUUUAAAGA--------GCUAAUUACAAG--- ---........(((.....)))......(((((..(((((((-----(.(((.....))).)))...))--------)))....)))))--- ( -12.80, z-score = -0.58, R) >droEre2.scaffold_4770 4458015 88 + 17746568 ---CAACUU-ACAGGUUUUCCCGCCACUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCUCCAUUCCCCUAUUUCAACGAA ---......-..(((.......((((....)))).(((((((((((((.......)))))))))))))..........)))........... ( -20.00, z-score = -2.24, R) >droYak2.chr3R 12649166 88 - 28832112 ---CAACUU-ACAGGUUUUCCCGCCAUUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCUCUAUUCCUCUAUUUCAACGAA ---......-..(((.((((..((((....)))).........(((((((.....)))))))))))..)))..................... ( -20.20, z-score = -2.33, R) >droSec1.super_0 7498610 85 + 21120651 CUGCAACUU-ACAGGUUUUCCCGCCAUUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCU------CUCUAUUUCAGCGAA .(((.....-(((((.....))((((....))))......)))(((((((.....)))))))(((((....------....))))).))).. ( -22.00, z-score = -1.80, R) >droSim1.chr3R 14451679 85 + 27517382 CUGCAACUU-ACAGGUUUUCCCGCCAUUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCU------CUCUAUUUCAGCGAA .(((.....-(((((.....))((((....))))......)))(((((((.....)))))))(((((....------....))))).))).. ( -22.00, z-score = -1.80, R) >consensus ___CAACUU_ACAGGUUUUCCCGCCAUUUUUGGCCAGUUUUGUGUGUGCGCAUUCCGCACGCGAAAUUCCU______CUCUAUUUCAACGAA ............(((.((((..((((....)))).........(((((((.....)))))))))))..)))..................... (-12.93 = -13.90 + 0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:22 2011