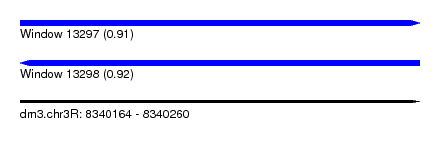
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,340,164 – 8,340,260 |
| Length | 96 |
| Max. P | 0.922166 |

| Location | 8,340,164 – 8,340,260 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.57426 |
| G+C content | 0.47607 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -14.37 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
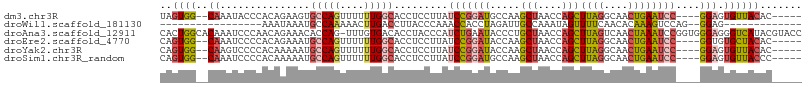
>dm3.chr3R 8340164 96 + 27905053 -----GUGUAACACUCC----GGAUUCAGUUGCCUAAGCUGGUUAGCUUGGCAUCCGGAUAAGGAGGUGCCAAAAAACUGGCACUUCUGUGGGUAUUUG--CCACUA -----......((((((----(((......((((.(((((....)))))))))))))))...((((((((((......)))))))))))))(((....)--)).... ( -39.80, z-score = -3.76, R) >droWil1.scaffold_181130 2136110 75 + 16660200 -------------CUCC--CUGGACUUUGUGUUGAAAACUAUUUGGCAAUCUAGGUGGUUUGGGUAAGGUCAAGUUUUUGGCAUUUAUUU----------------- -------------..((--..((((((..(.(((.(((((((((((....)))))))))))...))).)..))))))..)).........----------------- ( -16.30, z-score = -0.41, R) >droAna3.scaffold_12911 3271388 106 + 5364042 GGUACGUAUGACCCUCCCACCGGAUUUAGUUGACUAAGCUGGUUAGCAGGGUAUUCAGAUGGGUAGGUGUCACAAA-CUGGUGUUUCUGUUGGGAUUUGUGCCAGUG ........((((((((((((.((((...(((((((.....))))))).....)))).).)))).))).))))...(-((((..(.(((....)))...)..))))). ( -32.60, z-score = -0.46, R) >droEre2.scaffold_4770 4433963 96 - 17746568 -----GUGUAGCACACC----GGAUUCAGUUGCCUAAGCUGGUUAGCUUGGUAUCCGGAUAAGGAGGUGCCAAAAAACUGGCAUUUCUGUGGGGAUUUG--CCACUG -----(((...))).((----(((((((((((((......)).)))).))).))))))....((((((((((......))))))))))((((.......--)))).. ( -34.30, z-score = -1.92, R) >droYak2.chr3R 12624139 96 + 28832112 -----GUGUAACACUCC----GGAUUCAGUUGCCUAAGCUGGUUAGCUUGGUAUCCGGAUAAGGAGGUGCCAAAAAACUGGCAUUUUUGUGGGGACUUG--CCACUG -----.........(((----(((((((((((((......)).)))).))).)))))))...((((((((((......))))))))))((((.......--)))).. ( -32.30, z-score = -1.66, R) >droSim1.chr3R_random 521735 96 + 1307089 -----GGGUAACACUCC----GGAUUCAGUUGCCUAAGCUGGUUAGCUUGGCAUCCGGAUAAGGAGGUGCCAAAAAACUGGCAUUUUUGUGGGGAUUUG--CCACUG -----.(((((((((((----(((......((((.(((((....)))))))))))))))...((((((((((......))))))))))))).....)))--)).... ( -35.40, z-score = -2.23, R) >consensus _____GUGUAACACUCC____GGAUUCAGUUGCCUAAGCUGGUUAGCUUGGUAUCCGGAUAAGGAGGUGCCAAAAAACUGGCAUUUCUGUGGGGAUUUG__CCACUG .....................((((((.(.((.(((((((....))))))))).).))))..((((((((((......)))))))))).............)).... (-14.37 = -14.55 + 0.18)

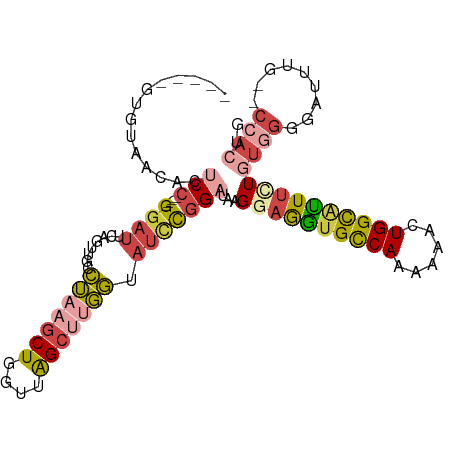
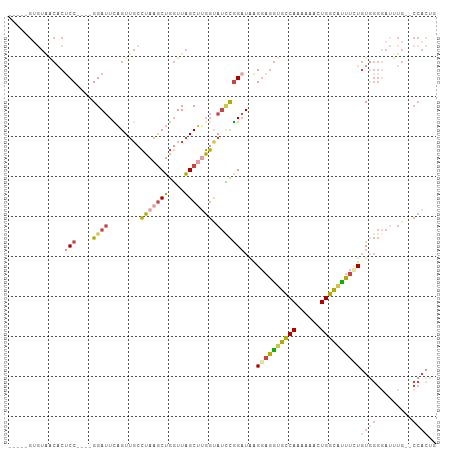
| Location | 8,340,164 – 8,340,260 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.57426 |
| G+C content | 0.47607 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -8.64 |
| Energy contribution | -9.25 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8340164 96 - 27905053 UAGUGG--CAAAUACCCACAGAAGUGCCAGUUUUUUGGCACCUCCUUAUCCGGAUGCCAAGCUAACCAGCUUAGGCAACUGAAUCC----GGAGUGUUACAC----- ..((((--((..........((.(((((((....))))))).))....(((((((((((((((....))))).))))......)))----))).))))))..----- ( -36.90, z-score = -4.56, R) >droWil1.scaffold_181130 2136110 75 - 16660200 -----------------AAAUAAAUGCCAAAAACUUGACCUUACCCAAACCACCUAGAUUGCCAAAUAGUUUUCAACACAAAGUCCAG--GGAG------------- -----------------..........................(((..........((..((......))..)).((.....))...)--))..------------- ( -5.10, z-score = 1.16, R) >droAna3.scaffold_12911 3271388 106 - 5364042 CACUGGCACAAAUCCCAACAGAAACACCAG-UUUGUGACACCUACCCAUCUGAAUACCCUGCUAACCAGCUUAGUCAACUAAAUCCGGUGGGAGGGUCAUACGUACC .(((((....................))))-).((((((.(((.((((((.((.......(((....)))((((....)))).)).)))))))))))))))...... ( -28.25, z-score = -1.74, R) >droEre2.scaffold_4770 4433963 96 + 17746568 CAGUGG--CAAAUCCCCACAGAAAUGCCAGUUUUUUGGCACCUCCUUAUCCGGAUACCAAGCUAACCAGCUUAGGCAACUGAAUCC----GGUGUGCUACAC----- ..((((--((..........((..((((((....))))))..)).....((((((.....(((....)))((((....))))))))----))..))))))..----- ( -31.40, z-score = -3.17, R) >droYak2.chr3R 12624139 96 - 28832112 CAGUGG--CAAGUCCCCACAAAAAUGCCAGUUUUUUGGCACCUCCUUAUCCGGAUACCAAGCUAACCAGCUUAGGCAACUGAAUCC----GGAGUGUUACAC----- ..((((--((..............((((((....))))))........(((((((.....(((....)))((((....))))))))----))).))))))..----- ( -30.50, z-score = -3.14, R) >droSim1.chr3R_random 521735 96 - 1307089 CAGUGG--CAAAUCCCCACAAAAAUGCCAGUUUUUUGGCACCUCCUUAUCCGGAUGCCAAGCUAACCAGCUUAGGCAACUGAAUCC----GGAGUGUUACCC----- ..((((--((..............((((((....)))))).((((..((.(((.(((((((((....))))).)))).))).))..----))))))))))..----- ( -32.10, z-score = -3.46, R) >consensus CAGUGG__CAAAUCCCCACAGAAAUGCCAGUUUUUUGGCACCUCCUUAUCCGGAUACCAAGCUAACCAGCUUAGGCAACUGAAUCC____GGAGUGUUACAC_____ .........................(((((....)))))..((((...............(((....)))((((....))))........))))............. ( -8.64 = -9.25 + 0.61)
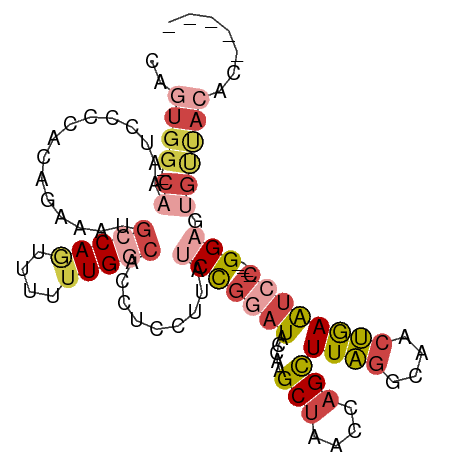
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:18 2011