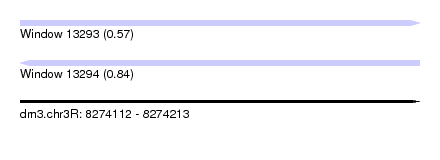
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,274,112 – 8,274,213 |
| Length | 101 |
| Max. P | 0.838775 |

| Location | 8,274,112 – 8,274,213 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 66.55 |
| Shannon entropy | 0.53059 |
| G+C content | 0.44159 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -10.40 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
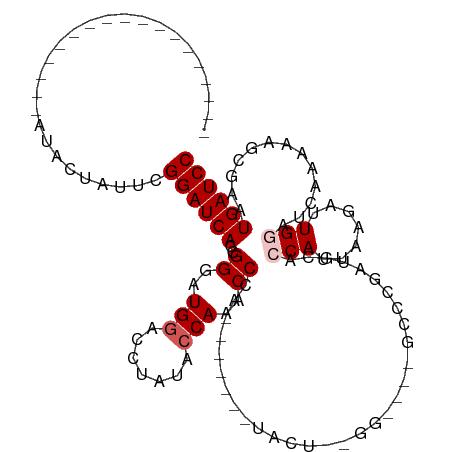
>dm3.chr3R 8274112 101 + 27905053 ------------------AUACUAUUCGGAUCACCGGGAUGGACCUAUACCAAACCCAUUCAAAGUACUUUGGUGUUGCCCGAUGCACCAUUAAGAUUGGAUCAAAAAGCGAAUGAUCC ------------------.((((...(((....)))((((((.............))))))..))))(((((((((........))))))..)))...((((((.........)))))) ( -23.52, z-score = -0.81, R) >droEre2.scaffold_4770 4399613 113 - 17746568 ACCACAAGUGGUAAGCAAAUAGAAUUCGGAUCACCGGGAUGGAAGUAUACCAUUCCAA------AUACUCCGGCUACGCCCCAAGCCCCAAUGAGCUUGAAUCAAAAGACGAAUGAUCC ((((....))))...............((((((..(((((((.......)))))))..------.......(((...))).(((((........)))))..............)))))) ( -27.80, z-score = -1.67, R) >droYak2.chr3R 12597443 119 + 28832112 ACCACAAAUAGUAAGCAAAUACUAUUCGGAUCACCGGGAUGGACGUAUACCAAACCCAUUCAAAGUACUCCGGCUACGCCCAGUGCAACAAUCAAAUUGAAUCAAAAAACGAAUGAUCC ......(((((((......))))))).(((((...(((.(((.......)))..)))((((...(((((..(((...))).)))))..((((...))))...........))))))))) ( -25.50, z-score = -2.27, R) >droSec1.super_0 7447392 77 - 21120651 ------------------AUACUAUUCGGAUCACCGGGAUGGUCCUAUACCAAACCC------------------------GAUGCACCAUUAAGAUUGGAUCAAAAAGCGAAUGAUCC ------------------.........((((((.((((.((((.....))))..)))------------------------).....(((.......))).............)))))) ( -18.70, z-score = -1.38, R) >droSim1.chr3R 14402022 77 - 27517382 ------------------AUACUAUUCGGAUCACCGGGAUGGUCCUAUAGCAAACCC------------------------GAUGCACCAUUAAGAUUGGAUCAAAAAGCGAAUGAUCC ------------------.........((((((.((((...((......))...)))------------------------).....(((.......))).............)))))) ( -16.30, z-score = -0.47, R) >consensus __________________AUACUAUUCGGAUCACCGGGAUGGACCUAUACCAAACCCA_______UACU__GG____GCCCGAUGCACCAUUAAGAUUGGAUCAAAAAGCGAAUGAUCC ...........................((((((..((..(((.......)))..))...............................(((.......))).............)))))) (-10.40 = -11.00 + 0.60)

| Location | 8,274,112 – 8,274,213 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.55 |
| Shannon entropy | 0.53059 |
| G+C content | 0.44159 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -14.46 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
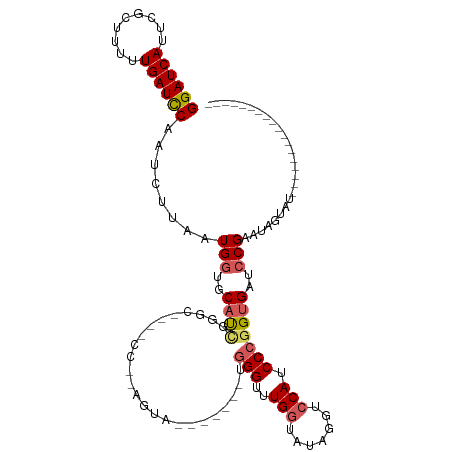
>dm3.chr3R 8274112 101 - 27905053 GGAUCAUUCGCUUUUUGAUCCAAUCUUAAUGGUGCAUCGGGCAACACCAAAGUACUUUGAAUGGGUUUGGUAUAGGUCCAUCCCGGUGAUCCGAAUAGUAU------------------ ((((((.........))))))........(((..(((((((......((((....)))).((((..(((...)))..)))))))))))..)))........------------------ ( -26.80, z-score = -0.58, R) >droEre2.scaffold_4770 4399613 113 + 17746568 GGAUCAUUCGUCUUUUGAUUCAAGCUCAUUGGGGCUUGGGGCGUAGCCGGAGUAU------UUGGAAUGGUAUACUUCCAUCCCGGUGAUCCGAAUUCUAUUUGCUUACCACUUGUGGU (((((...(((((.......(((((((....))))))))))))..(((((.(...------.(((((.(.....)))))).)))))))))))...............((((....)))) ( -35.91, z-score = -1.42, R) >droYak2.chr3R 12597443 119 - 28832112 GGAUCAUUCGUUUUUUGAUUCAAUUUGAUUGUUGCACUGGGCGUAGCCGGAGUACUUUGAAUGGGUUUGGUAUACGUCCAUCCCGGUGAUCCGAAUAGUAUUUGCUUACUAUUUGUGGU ((((((.........)))))).....(((..(((...((((((((((((((...((......)).)))))).))))))))...)))..)))(((((((((......))))))))).... ( -35.30, z-score = -2.01, R) >droSec1.super_0 7447392 77 + 21120651 GGAUCAUUCGCUUUUUGAUCCAAUCUUAAUGGUGCAUC------------------------GGGUUUGGUAUAGGACCAUCCCGGUGAUCCGAAUAGUAU------------------ ((((((.........))))))........(((..((((------------------------(((..((((.....)))).)))))))..)))........------------------ ( -26.70, z-score = -2.60, R) >droSim1.chr3R 14402022 77 + 27517382 GGAUCAUUCGCUUUUUGAUCCAAUCUUAAUGGUGCAUC------------------------GGGUUUGCUAUAGGACCAUCCCGGUGAUCCGAAUAGUAU------------------ ((((((.........))))))........(((..((((------------------------(((....(.....).....)))))))..)))........------------------ ( -21.20, z-score = -1.03, R) >consensus GGAUCAUUCGCUUUUUGAUCCAAUCUUAAUGGUGCAUCGGGC____CC__AGUA_______UGGGUUUGGUAUAGGUCCAUCCCGGUGAUCCGAAUAGUAU__________________ ((((((((.((...((((.......))))....))............................((..(((.......)))..))))))))))........................... (-14.46 = -14.90 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:14 2011