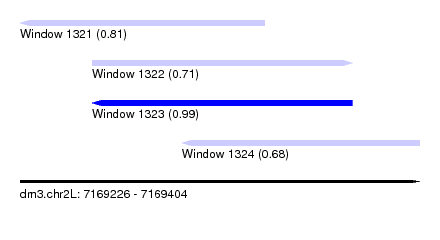
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,169,226 – 7,169,404 |
| Length | 178 |
| Max. P | 0.989379 |

| Location | 7,169,226 – 7,169,335 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.82 |
| Shannon entropy | 0.37441 |
| G+C content | 0.42442 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7169226 109 - 23011544 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUGUAUAAAGUGGGUGGCAAAA------- .......(((((..(((........--)))..))-))).(((((((....)))))))..((....))..(((((..(((....(((.....)))....)))..)))))....------- ( -30.30, z-score = -2.09, R) >droAna3.scaffold_12943 1019427 105 + 5039921 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGC-AGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUGUA-AACGAGGGGGACAA--------- ((((((((((((..(((........--)))..))-)))-.)))))))...((((.(((......))).))))(((..(((....)))..)))...-......(....)..--------- ( -28.10, z-score = -2.07, R) >droEre2.scaffold_4929 16084631 109 - 26641161 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGGCUGUGUAUAAAGUGGGUGGCCAAA------- ((((((((((((..(((........--)))..))-)))..)))))))...((((.(((......))).))))..............(((..(.(((...))).)..)))...------- ( -31.50, z-score = -1.52, R) >droYak2.chr2L 16588926 109 + 22324452 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACGGACUGUGUAUAAAGUGGGUGGCACAA------- .......(((((..(((........--)))..))-))).(((((((....)))))))..((....))..(((((..(((....(((.....)))....)))..)))))....------- ( -29.70, z-score = -1.55, R) >droSec1.super_3 2692709 109 - 7220098 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUGUAUAAAGUGGGUGGCAAAA------- .......(((((..(((........--)))..))-))).(((((((....)))))))..((....))..(((((..(((....(((.....)))....)))..)))))....------- ( -30.30, z-score = -2.09, R) >droSim1.chr2L 6958960 109 - 22036055 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUGUAUAAAGUGGGUGGUAAAA------- .......(((((..(((........--)))..))-))).(((((((....)))))))..((....))..(((((..(((....(((.....)))....)))..)))))....------- ( -28.00, z-score = -1.58, R) >droWil1.scaffold_180772 3257886 103 + 8906247 UAAUUGCGCCAUAAAUCAAAUCUAGGAGAUAAAUGGGCCAACAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAAGAGAGAGAGAGCAACUGAC---------------- ...(((((((....(((..........))).....)))...(((((....((((.(((......))).))))...)))))..............)))).....---------------- ( -22.20, z-score = -0.91, R) >droMoj3.scaffold_6500 4363823 88 - 32352404 UAAUUGCCCCAUAAAUCAAAUCUAG--GAUAAAU-GGCCAGCAGUUAAAGGGCUGCUAACCAAAUGGCAGCCAUAAGCUGAUAACAGACUC---------------------------- (((((((.((((..(((........--)))..))-))...)))))))...((((((((......))))))))...(((((....))).)).---------------------------- ( -25.60, z-score = -2.55, R) >droGri2.scaffold_15252 15678837 104 - 17193109 UAAUUGCGCCAUAAAUCAAAUCUAG--GAUAAAU-GGCCGGCAGUUAAAGGGCUGCUAACGAAAUGGCAGCCAUAAGCUGAUAACAAAACUCAAGGGGGCAGUCAGA------------ ((((((((((((..(((........--)))..))-)))..)))))))...((((((((......)))))))).....(((((..(....((....)).)..))))).------------ ( -32.10, z-score = -2.78, R) >droPer1.super_8 873553 116 + 3966273 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUCUAAAGGGGGGGGGAUGUGGGGUAGCA ((((((((((((..(((........--)))..))-)))..)))))))...((((.(((......))).))))....((((...(((..((.(((.....))).))..)))....)))). ( -36.20, z-score = -2.67, R) >dp4.chr4_group2 809015 114 - 1235136 UAAUUGCGCCAUAAAUCAAAUCUAC--GAUAAAU-GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUCUAAAG--GGGGGGAUGUGGGGUAGCA ((((((((((((..(((........--)))..))-)))..)))))))...((((.(((......))).))))....((((...(((..((.((.....--)).))..)))....)))). ( -35.30, z-score = -2.49, R) >consensus UAAUUGCGCCAUAAAUCAAAUCUAC__GAUAAAU_GGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUGAUAACAGACUGUGUAUAAAGUGGGUGGCAAAA_______ ((((((((((((..(((..........)))..)).)))..)))))))...((((.(((......))).)))).....(((....)))................................ (-16.89 = -17.17 + 0.28)
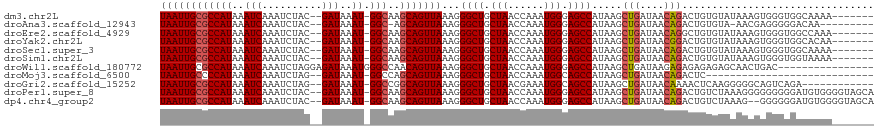
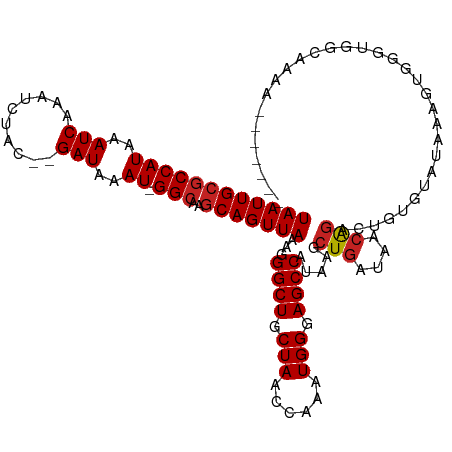
| Location | 7,169,258 – 7,169,374 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Shannon entropy | 0.15344 |
| G+C content | 0.45438 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7169258 116 + 23011544 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG-GGGGGGGGGUUGGAGU .((((.((((...))))..))))..(((((((((((.(((..(((((..((((......))))..)))))))).)))))..((.(((((..(....).))-))).)))))))).... ( -40.40, z-score = -1.50, R) >droAna3.scaffold_12943 1019456 108 - 5039921 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCU-GCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAAGACGGCGAUGG-------- ..(((.........(((((((...((((...(((((.(((.-(((((..((((......))))..)))))))).)))))..)))).)))))))........))).....-------- ( -31.33, z-score = -1.96, R) >droEre2.scaffold_4929 16084663 111 + 26641161 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG-GGCGGGGCUUG----- ........(((((((((((((...((((...(((((.(((..(((((..((((......))))..)))))))).)))))..)))).)))))))....(..-..)))))))..----- ( -35.40, z-score = -1.36, R) >droYak2.chr2L 16588958 112 - 22324452 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG-GGCGGGGUGUUA---- .((((.((((...))))..))))((((.((((((((.(((..(((((..((((......))))..)))))))).)))))....((((....(....)..)-)))))).)))).---- ( -35.10, z-score = -1.27, R) >droSec1.super_3 2692741 111 + 7220098 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG-GGCGGAGGUUU----- ..((((((......(((((((...((((...(((((.(((..(((((..((((......))))..)))))))).)))))..)))).)))))))....)))-)))........----- ( -34.00, z-score = -1.42, R) >droSim1.chr2L 6958992 111 + 22036055 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG-GGCGGGGGUUU----- ........(((((((.((((((((((((........))))).(((((..((((......))))..)))))..)))))))....((((....(....)..)-)))))))))).----- ( -35.00, z-score = -1.11, R) >droPer1.super_8 873592 110 - 3966273 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAGGGGA-GGAGCCGUUC------ ......(((((((((((((((...((((...(((((.(((..(((((..((((......))))..)))))))).)))))..)))).))))))).......-))))))))..------ ( -39.31, z-score = -3.11, R) >dp4.chr4_group2 809052 110 + 1235136 CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAGGGGA-GGAGCGGUUC------ ..((((....(((((((((((...((((...(((((.(((..(((((..((((......))))..)))))))).)))))..)))).)))))))..)))).-.)))).....------ ( -38.10, z-score = -2.48, R) >consensus CAGCUUAUGGCUCCCAUUUGGUUAGCAGCCCUUUAACUGCUUGCCAUUUAUCGUAGAUUUGAUUUAUGGCGCAAUUAAAAACUGCCCCAGAUGAAAAGUG_GGCGGGGGUUU_____ .(((.....)))..(((((((...((((...(((((.(((..(((((..((((......))))..)))))))).)))))..)))).)))))))........................ (-29.80 = -29.80 + 0.00)


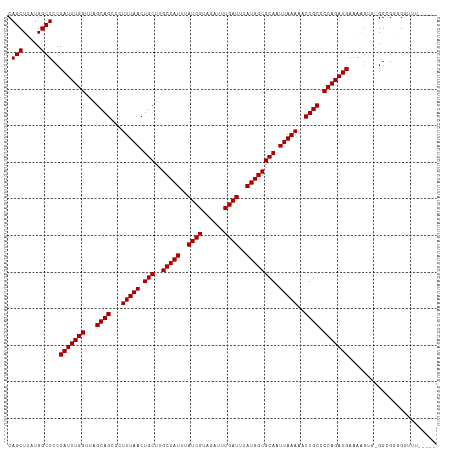
| Location | 7,169,258 – 7,169,374 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Shannon entropy | 0.15344 |
| G+C content | 0.45438 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.27 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
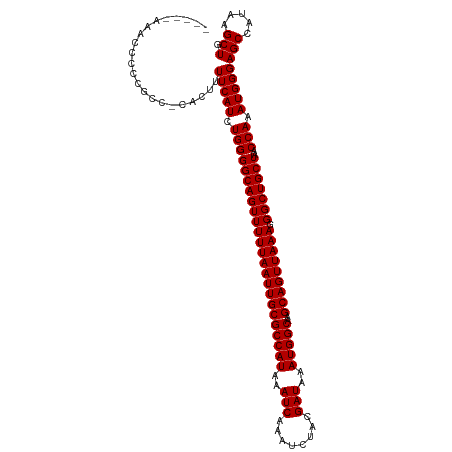
>dm3.chr2L 7169258 116 - 23011544 ACUCCAACCCCCCCCC-CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG .............(((-((.........)))))(((((((((((((((((((..(((........)))..)))))..))))))))).((((.(((......))).))))...))))) ( -34.50, z-score = -2.16, R) >droAna3.scaffold_12943 1019456 108 + 5039921 --------CCAUCGCCGUCUUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGC-AGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG --------.....((.....((..(((.((((((((((((((((((((((((..(((........)))..)))))-.))))))))).)))))))..))).)))..))......)).. ( -34.90, z-score = -2.79, R) >droEre2.scaffold_4929 16084663 111 - 26641161 -----CAAGCCCCGCC-CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG -----...((((((..-...........))))))((((((((((((((((((..(((........)))..)))))..))))))))).((((.(((......))).))))...)))). ( -36.22, z-score = -2.35, R) >droYak2.chr2L 16588958 112 + 22324452 ----UAACACCCCGCC-CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG ----......((((((-((.........))))((((((((((((((((((((..(((........)))..)))))..))))))))).))))))........))))(((.....))). ( -34.40, z-score = -2.35, R) >droSec1.super_3 2692741 111 - 7220098 -----AAACCUCCGCC-CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG -----....((((.((-((.........))))((((((((((((((((((((..(((........)))..)))))..))))))))).))))))..........)))).......... ( -33.40, z-score = -2.10, R) >droSim1.chr2L 6958992 111 - 22036055 -----AAACCCCCGCC-CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG -----.....((((((-((.........))))((((((((((((((((((((..(((........)))..)))))..))))))))).))))))........))))(((.....))). ( -34.40, z-score = -2.13, R) >droPer1.super_8 873592 110 + 3966273 ------GAACGGCUCC-UCCCCUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG ------....((((((-(.(((.......)))((((((((((((((((((((..(((........)))..)))))..))))))))).)))))).........)))))))........ ( -41.90, z-score = -4.29, R) >dp4.chr4_group2 809052 110 - 1235136 ------GAACCGCUCC-UCCCCUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG ------.....(((((-(.(((.......)))((((((((((((((((((((..(((........)))..)))))..))))))))).)))))).........))))))......... ( -37.70, z-score = -3.31, R) >consensus _____AAACCCCCGCC_CACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAAGCAGUUAAAGGGCUGCUAACCAAAUGGGAGCCAUAAGCUG ......................(((((.((((((((((((((((((((((((..(((........)))..)))))..))))))))).)))))))..))).)))))(((.....))). (-32.28 = -32.27 + -0.00)

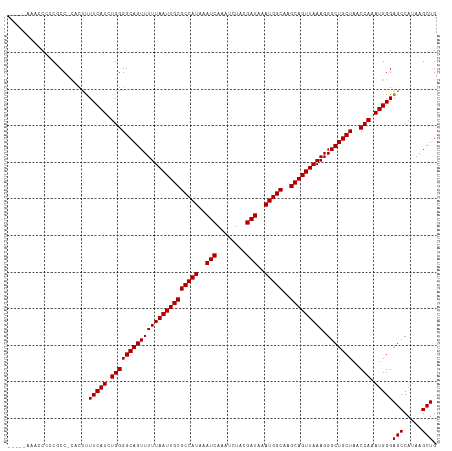
| Location | 7,169,298 – 7,169,404 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.83 |
| Shannon entropy | 0.40672 |
| G+C content | 0.46271 |
| Mean single sequence MFE | -19.45 |
| Consensus MFE | -14.01 |
| Energy contribution | -13.90 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
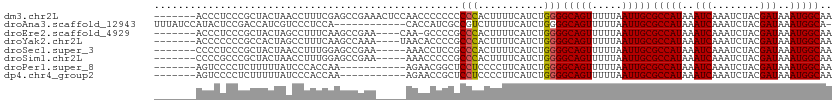
>dm3.chr2L 7169298 106 - 23011544 -------ACCCUCCCGCUACUAACCUUUCGAGCCGAAACUCCAACCCCCCCCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------........((............(((......))).........(((((.........)))))............))(((((..(((........)))..))))).. ( -18.30, z-score = -1.31, R) >droAna3.scaffold_12943 1019496 100 + 5039921 UUUAUCCAUACUCCGACCAUCGUCCCUCCA------------CACCAUCGCCGUCUUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCA- ..............(((....)))......------------.......(((((..........(((.(((((.....))))).)))...(((........)))..))))).- ( -17.20, z-score = -1.14, R) >droEre2.scaffold_4929 16084703 101 - 26641161 -------ACCCUCCCGCUACUAGCCUUUCAAGCCGAA----CAA-GCCCCGCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------........((.....((.......))....----...-((((((.............))))))...........))(((((..(((........)))..))))).. ( -20.02, z-score = -0.66, R) >droYak2.chr2L 16588998 102 + 22324452 -------ACCCCCCCGCCACUAGCCUUUCAAGCCAAA----UAACACCCCGCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------........((.....((.......))....----.........((((.(.........)))))...........))(((((..(((........)))..))))).. ( -16.70, z-score = -0.16, R) >droSec1.super_3 2692781 101 - 7220098 -------CCCCUCCCGCUACUAACCUUUGGAGCCGAA-----AAACCUCCGCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------.....((((...........(((.((.((.-----.....)).))))).........))))(((((.....)))))(((((..(((........)))..))))).. ( -21.55, z-score = -1.21, R) >droSim1.chr2L 6959032 101 - 22036055 -------CCCCGCCCGCUACUAACCUUUGGAGCCGAA-----AAACCCCCGCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------.(((....(((.(((.....)))))).(((-----((............)))))....)))(((((.....)))))(((((..(((........)))..))))).. ( -21.60, z-score = -0.65, R) >droPer1.super_8 873632 95 + 3966273 -------AGUCCCCUCUUUUUAUCCCACCAA-----------AGAACGGCUCCUCCCCUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------.......................(-----------(((((.(((((............))))).))))))......(((((..(((........)))..))))).. ( -20.10, z-score = -1.57, R) >dp4.chr4_group2 809092 95 - 1235136 -------AGUCCCCUCUUUUUAUCCCACCAA-----------AGAACCGCUCCUCCCCUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA -------.......................(-----------(((((.(((((............))))).))))))......(((((..(((........)))..))))).. ( -20.10, z-score = -2.35, R) >consensus _______ACCCUCCCGCUACUAACCUUUCAAGCCGAA_____AAACCCCCGCCCACUUUUCAUCUGGGGCAGUUUUUAAUUGCGCCAUAAAUCAAAUCUACGAUAAAUGGCAA ...................................................(((...........)))(((((.....)))))(((((..(((........)))..))))).. (-14.01 = -13.90 + -0.11)
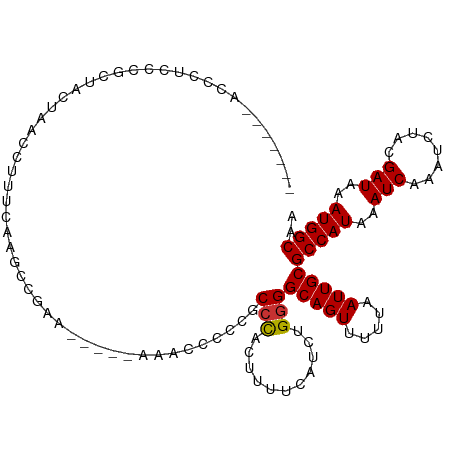
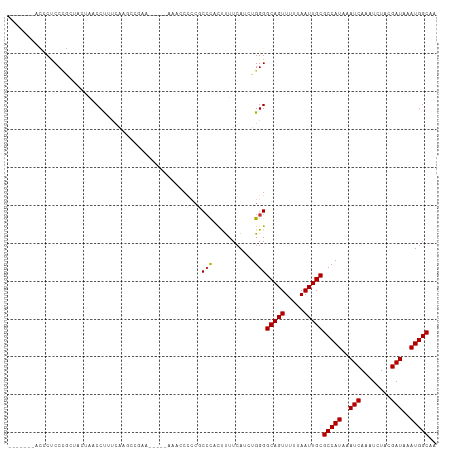
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:07 2011