
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,271,309 – 8,271,402 |
| Length | 93 |
| Max. P | 0.634212 |

| Location | 8,271,309 – 8,271,402 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 68.90 |
| Shannon entropy | 0.57677 |
| G+C content | 0.48462 |
| Mean single sequence MFE | -31.69 |
| Consensus MFE | -4.95 |
| Energy contribution | -4.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8271309 93 - 27905053 AACUUUGUGAAGGUUAUCAG-GG------CGCAAGCCUUA--------GAUAACCCCUAUAUGUUCGAGAGUUCGGCACGCGACAGGAAGAUGGCAAUGGUUGUCAUG ..((((.((..(((((((((-((------(....))))).--------))))))).......(((((......))).))....)).))))((((((.....)))))). ( -31.50, z-score = -2.62, R) >dp4.chr2 5498896 98 - 30794189 CACUUAACGAAGGUUAUCAGUGCUGGACUCGUAUGGAUCACAAACACUGAUAGCAUUGGUAUAUUCAAGCGUCCAGCACGCGCUAGAAAGAUGACCAU---------- ...........(((((((.(((((((((..((.((.....)).)).......((.((((.....))))))))))))))).(....)...)))))))..---------- ( -32.00, z-score = -3.56, R) >droPer1.super_19 1215393 98 - 1869541 CACUUAACGAAGGUUAUCAGUGCUGGACUCGUAUGGAUCACAAACACUGAUAGCUUUGGUAUAUUCAAGCGUCCGGCACGCGCUGGAAAGAUGACCAU---------- ......((.(((((((((((((.((((..(....)..)).))..))))))))))))).))....(((....((((((....))))))....)))....---------- ( -31.60, z-score = -2.83, R) >droSim1.chr3R 14399173 93 + 27517382 AACUUUGUGAAGGUUAUCAG-GG------CGCAAGCCUUA--------GAUAACCCCUAUAUGUUCGAGGGUUCGGCACGCGACAGGAAGAUGGCAAUGGUUGUCAUG ..((((.((..(((((((((-((------(....))))).--------))))))).......(((((......))).))....)).))))((((((.....)))))). ( -31.50, z-score = -2.26, R) >droSec1.super_0 7444544 93 + 21120651 AACUUUGUGAAGGUUAUCAG-GG------CGCAAGCCUUA--------GAUAACCCCUAUAUGUUCGAGGGUUCGGCACGCGACAGGAAGAUGGCAAUGGUUGCCAUG ..((((.((..(((((((((-((------(....))))).--------))))))).......(((((......))).))....)).))))((((((.....)))))). ( -34.20, z-score = -2.78, R) >droYak2.chr3R 12594664 93 - 28832112 AACUUGGUGAAGGUUAUCAG-GG------CGCAAGCCUUA--------GAUAACCUCUAUAUGGUCGAGAGUUCGGCACGCGACAGGAAGAUGGCAAUGGCUGUCAAG ..(((((((.((((((((((-((------(....))))).--------)))))))).)))...((((......))))......))))..((((((....))))))... ( -37.00, z-score = -4.13, R) >droEre2.scaffold_4770 4396782 89 + 17746568 ----AACUGAAGGUUAUCAG-GG------CGCAAGCCUUA--------GAUAACCCCUAUAUGGUCGAGAGUUCGGCACGCGACAGGAAGAUGACAAUGGUUGUCAAG ----..(((..(((((((((-((------(....))))).--------)))))))........((((......))))......)))...((..((....))..))... ( -32.10, z-score = -3.61, R) >droAna3.scaffold_13340 7652753 80 - 23697760 GACUAUGUGAAGGUUAUCAGUGC------CUUGGAAAUUA--------GAUAACCCUCA--------------CGGCACGCGCCAGGAAGAUGAAAAUCGUUGCCAUG ......((((.((((((((((..------.......))).--------))))))).)))--------------)(((....))).((..(((....)))....))... ( -23.60, z-score = -2.31, R) >consensus AACUUUGUGAAGGUUAUCAG_GG______CGCAAGCCUUA________GAUAACCCCUAUAUGUUCGAGAGUUCGGCACGCGACAGGAAGAUGACAAUGGUUGUCAUG ...........(((((((..............................)))))))..................................................... ( -4.95 = -4.80 + -0.16)

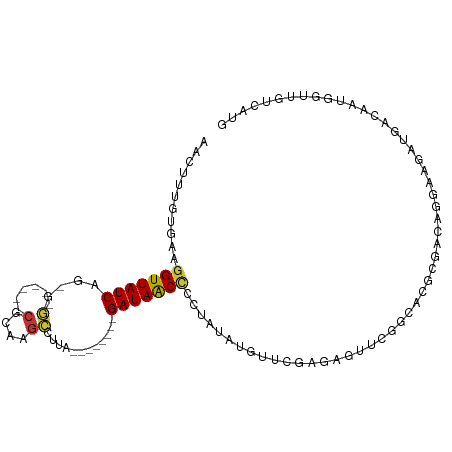

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:13 2011