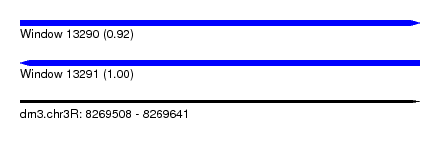
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,269,508 – 8,269,641 |
| Length | 133 |
| Max. P | 0.997126 |

| Location | 8,269,508 – 8,269,641 |
|---|---|
| Length | 133 |
| Sequences | 6 |
| Columns | 136 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Shannon entropy | 0.38826 |
| G+C content | 0.44632 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.61 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
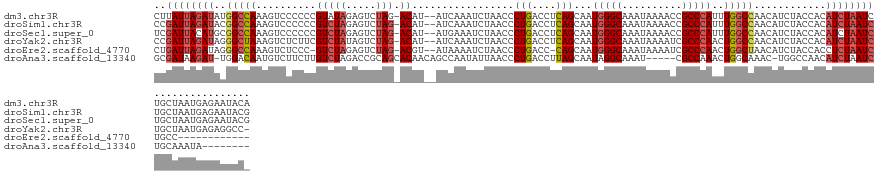
>dm3.chr3R 8269508 133 + 27905053 CUUAUUAGAUAUGGCCAAAGUCCCCCCGUAUAGAGUCUAG-ACAU--AUCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACA ((((((((...((((((((.............(((..(((-....--.............)))..))).....(((((..........)))))))))))))..........((.......))))))))))...... ( -28.33, z-score = -2.36, R) >droSim1.chr3R 14397406 133 - 27517382 CCGAUUAGAUACGGCCAAAGUCCCCCCGUCUAGAGUCUAG-ACAU--AUCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACG ..((((((((..(((((((........((((((...))))-))..--.............(((....)))...(((((..........))))))))))))............))))))))................ ( -31.04, z-score = -3.40, R) >droSec1.super_0 7442769 133 - 21120651 UCGAUUACAUGCGGCCAAAGUCCCCCCGUCUAGAGUCUAG-ACAU--AUGAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAUUUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUACG ..(((((.(((.(((((((........((((((...))))-))..--.............(((....)))...(((((..........))))))))))))...........))).)))))................ ( -27.80, z-score = -1.86, R) >droYak2.chr3R 12592908 132 + 28832112 CCGAUUAGAUAGGGCUAAAGUCUCUUCGUCUAUAGUCUAG-ACAU--AUCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUAAAAUCGCCCAACUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAGGCC- ..((((.((((((((....))))....(((((.....)))-)).)--))).)))).........((((((((.(((((..........)))))..(((..........))).........)))).....))))..- ( -27.80, z-score = -0.78, R) >droEre2.scaffold_4770 4395124 119 - 17746568 CUGAUUAGAUAGGGCCAAAGUCUCCC-GUCUAGAGUCUAG-ACGU--AUAAAAUCUAACCCUGACC-CAGCAAUGGGCAAAUAAAAUCGCCCAACUGGCUAACAUCUACCACCUCUAAUCUGCC------------ ..((((((((((((...........(-((((((...))))-))).--...........)))))..(-(((...(((((..........))))).))))...............)))))))....------------ ( -32.10, z-score = -3.41, R) >droAna3.scaffold_13340 7651433 121 + 23697760 GCGAUAAGAU-UGGACAAUGUCUUCUUGUCUAGACCGCAGCACAACAGCCAAUAUUAACCCUGACCUUAGCAAUAGGCAAAU-----CGCCAAACUGGCAAAC-UGGCCAACAUCUAAUCUGCAAAUA-------- ..(((((((.-.((((...)))))))))))......((((.......((((..........((.(((.......)))))...-----.(((.....)))....-))))...........)))).....-------- ( -25.27, z-score = -0.80, R) >consensus CCGAUUAGAUACGGCCAAAGUCCCCCCGUCUAGAGUCUAG_ACAU__AUCAAAUCUAACCCUGACCUCAGCAAUGGGCAAAUAAAACCGCCCAACUGGCCAACAUCUACCACAUCUAAUCUGCUAAUGAGAAUAC_ ..((((((((..(((((...........................................(((....)))...(((((..........)))))..)))))............))))))))................ (-16.83 = -17.61 + 0.78)


| Location | 8,269,508 – 8,269,641 |
|---|---|
| Length | 133 |
| Sequences | 6 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Shannon entropy | 0.38826 |
| G+C content | 0.44632 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -29.86 |
| Energy contribution | -31.39 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8269508 133 - 27905053 UGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUUUAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGAU--AUGU-CUAGACUCUAUACGGGGGGACUUUGGCCAUAUCUAAUAAG .................((((((((((((.(((.(..((..(((((((((......)))))))))))..).)))((((((((((.....--..))-)).))))))....(((....)))..))))))))))))... ( -46.80, z-score = -3.53, R) >droSim1.chr3R 14397406 133 + 27517382 CGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUUUAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGAU--AUGU-CUAGACUCUAGACGGGGGGACUUUGGCCGUAUCUAAUCGG ................(((((((((((((.(((.(..((..(((((((((......)))))))))))..).)))((((((((((.....--..))-)).))))))....(((....)))..))))))))))))).. ( -47.10, z-score = -3.18, R) >droSec1.super_0 7442769 133 + 21120651 CGUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUUUAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUCAU--AUGU-CUAGACUCUAGACGGGGGGACUUUGGCCGCAUGUAAUCGA ................(((((.((((....(((.(..((..(((((((((......)))))))))))..).)))..(((((((.(((..--.(((-((((...)))))))...))).))))))))))).))))).. ( -48.30, z-score = -3.40, R) >droYak2.chr3R 12592908 132 - 28832112 -GGCCUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAGUUGGGCGAUUUUAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGAU--AUGU-CUAGACUAUAGACGAAGAGACUUUAGCCCUAUCUAAUCGG -...............((((((((......(((.(..((....(((((((......)))))))..))..).)))(((((((((.((..(--.(((-(((.....)))))).)..)).))))))))))))))))).. ( -47.30, z-score = -3.42, R) >droEre2.scaffold_4770 4395124 119 + 17746568 ------------GGCAGAUUAGAGGUGGUAGAUGUUAGCCAGUUGGGCGAUUUUAUUUGCCCAUUGCUG-GGUCAGGGUUAGAUUUUAU--ACGU-CUAGACUCUAGAC-GGGAGACUUUGGCCCUAUCUAAUCAG ------------....(((((((...............((((((((((((......)))))))..))))-)...(((((((((.(((..--.(((-((((...))))))-)..))).))))))))).))))))).. ( -47.60, z-score = -4.48, R) >droAna3.scaffold_13340 7651433 121 - 23697760 --------UAUUUGCAGAUUAGAUGUUGGCCA-GUUUGCCAGUUUGGCG-----AUUUGCCUAUUGCUAAGGUCAGGGUUAAUAUUGGCUGUUGUGCUGCGGUCUAGACAAGAAGACAUUGUCCA-AUCUUAUCGC --------.......(((((.((((((((((.-....(((...((((((-----((......)))))))))))...))))))))))((((((......))))))..(((((.......))))).)-))))...... ( -31.60, z-score = 0.17, R) >consensus _GUAUUCUCAUUAGCAGAUUAGAUGUGGUAGAUGUUGGCCAAAUGGGCGGUUUUAUUUGCCCAUUGCUGAGGUCAGGGUUAGAUUUGAU__AUGU_CUAGACUCUAGACGGGGAGACUUUGGCCAUAUCUAAUCGG ................(((((((((((((.(((.(((((.((.(((((((......)))))))))))))).)))((((((...................))))))................))))))))))))).. (-29.86 = -31.39 + 1.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:12 2011