
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,214,549 – 8,214,640 |
| Length | 91 |
| Max. P | 0.580539 |

| Location | 8,214,549 – 8,214,640 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.59961 |
| G+C content | 0.47432 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -12.34 |
| Energy contribution | -12.31 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
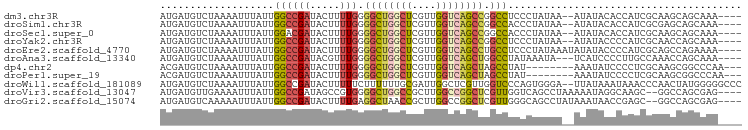
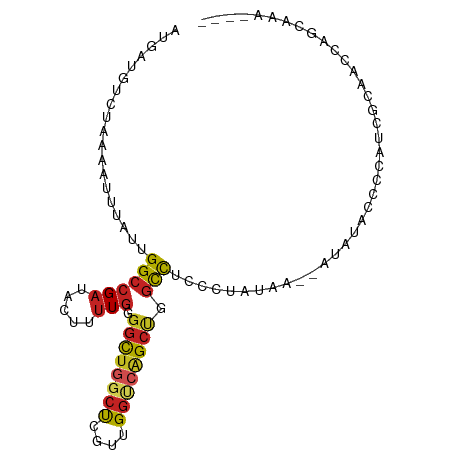
>dm3.chr3R 8214549 91 - 27905053 AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCCGGCCUCCCUAUAA--AUAUACACCAUCGCAAGCAGCAAA---- ..((((......((((((.((((((.....))))((((((((........))))))))..)).))))--))......))))((.....))...---- ( -26.50, z-score = -1.29, R) >droSim1.chr3R 14344443 91 + 27517382 AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCCGGCCACCCUAUAA--AUAUACACCAUCGCGAGCAGCAAA---- ..((((......((((((.((((((.....))))((((((((........))))))))..)).))))--))......))))((.....))...---- ( -27.70, z-score = -1.44, R) >droSec1.super_0 7384967 91 + 21120651 AUGAUGUCUAAAAUUUAUUGGACGAUACUUUUGGGGCUGGCUCGUUGGUCAGCCGGCCACCCUAUAA--AUAUACACCAUCGCAAGCAGCAAA---- ..(((((((((......)))))).(((.((.(((((.(((((.(((....))).)))))))))).))--.))).....)))((.....))...---- ( -24.90, z-score = -1.15, R) >droYak2.chr3R 12540524 91 - 28832112 AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCCGGCCUCCCUAUAA--AUAUACCCCAUCGCAACCAGCAAA---- ..((((......((((((.((((((.....))))((((((((........))))))))..)).))))--))......))))((.....))...---- ( -26.50, z-score = -1.39, R) >droEre2.scaffold_4770 4344399 93 + 17746568 AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCCUGCCUCCCUAUAAAUAUAUACCCCAUCGCAGCCAGAAAA---- ..((((......((((((.((((((.....))))((((((((....))))))))......)).))))))........))))............---- ( -21.94, z-score = -0.07, R) >droAna3.scaffold_13340 7602120 90 - 23697760 AUGAUGUCUAAAAUUUAUUGGCCGAUACGUUUGGGGCUGGCUCGUUGGUCAGCUGGCCUAUAAAUA---UCAUCCCCUUGCCAAACCAGCAAA---- ..((((......((((((.((((....(.....)((((((((....)))))))))))).)))))).---.))))...((((.......)))).---- ( -23.90, z-score = -0.41, R) >dp4.chr2 5444113 86 - 30794189 ACGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCUAGCCUAU--------AAAUAUCCCCUCGCAAGCGGCCCAA--- ...................((((((((.((...(((((((((........)))))))))..--------)).))))....(....).))))...--- ( -25.50, z-score = -0.92, R) >droPer1.super_19 1160326 86 - 1869541 ACGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCUAGCCUAU--------AAAUAUCCCCUCGCAAGCGGCCCAA--- ...................((((((((.((...(((((((((........)))))))))..--------)).))))....(....).))))...--- ( -25.50, z-score = -0.92, R) >droWil1.scaffold_181089 5359286 95 - 12369635 AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUUCUUUUUUGCGAUUGGCUCGUUGGUCCCAGUGGGA--UUAUAAAUAAACCCAACUAUGGGGGCCC ...................(((((((.................)))))))....((((((..((((.--(((....))).))))......)))))). ( -22.33, z-score = -0.09, R) >droVir3.scaffold_13047 5515230 91 + 19223366 AUGAUGUUGAAAAUUUAUUGGCCGAUAGCCGUGGGGCUGGCCGCUUGGCCGGCUCGUUGGUCAGCCUAAAAAUAGGCAAGC--GGCCAGCGAG---- ...................(((.....))).....(((((((((((((((((....)))))).(((((....)))))))))--)))))))...---- ( -41.30, z-score = -3.00, R) >droGri2.scaffold_15074 331133 91 - 7742996 AUGAUGUCAAAAAUUUAUUGGCCGAUACUUUUGAGGCUAACCGCUUGGCCGGCUCGUUGGGCAGCCUAUAAAUAACCGAGC--GGCCAGCGAG---- .................((((((...........)))))).((((.(((((.((((.((((...))))........)))))--))))))))..---- ( -28.40, z-score = -0.94, R) >consensus AUGAUGUCUAAAAUUUAUUGGCCGAUACUUUUGGGGCUGGCUCGUUGGUCAGCUGGCCUCCCUAUAA__AUAUACCCCAUCGCAACCAGCAAA____ ...................((((((.....))).((((((((....)))))))).)))....................................... (-12.34 = -12.31 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:07 2011