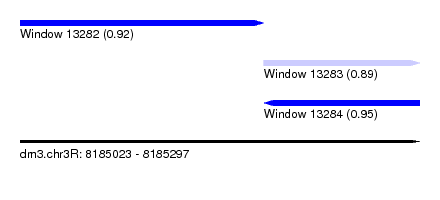
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,185,023 – 8,185,297 |
| Length | 274 |
| Max. P | 0.945766 |

| Location | 8,185,023 – 8,185,190 |
|---|---|
| Length | 167 |
| Sequences | 6 |
| Columns | 178 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Shannon entropy | 0.46719 |
| G+C content | 0.48934 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -21.94 |
| Energy contribution | -23.63 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
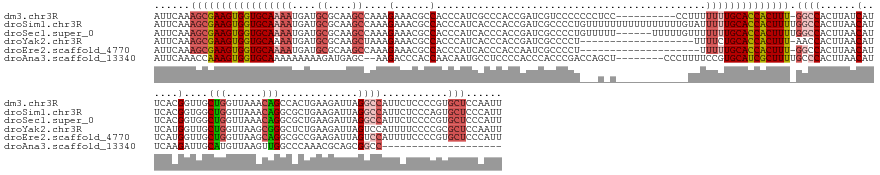
>dm3.chr3R 8185023 167 + 27905053 AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCCAAAGAAACGCCACCCAUCGCCCACCGAUCGUCCCCCCCUCC----------CCUUUUUUUGCACCACUUU-GGCCACUUAUCAUUCACGGUUGCUGGUUAAACAGCCACUGAAGAUUAGGCCAUUCUCCCCGUGCUCCAAUU ......(((((((((((((((((....((....))................((((.....)))).............----------.....))))))))))))))-((((....(((.((((..((.((((......)))).)))))))))..))))...........)))...... ( -46.10, z-score = -3.02, R) >droSim1.chr3R 14313496 178 - 27517382 AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCCAAAGAAACGCCACCCAUCACCCACCGAUCGCCCCUGUUUUUUUUUUUUUUUUGUAUUUUUGCACCACUUUUGGCCACUUAACAUUCACGGUGGCUGGUUAAACAGGCGCUGAAGAUUAGGCCAUUCUCCCAGUGCUCCCAUU ......(((((((((((((((((....((....)).(((((((((....(.(((.......))).)....))))))))).............))))))))))))))(((((...(((..((((..(((.(((......))).)))))))..))))))))..........)))...... ( -49.20, z-score = -1.65, R) >droSec1.super_0 7358029 172 - 21120651 AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCCAAAGAAACGCCACCCAUCACCCACCGAUCGCCCCUGUUUUU------UUUUUGUUUUUUUGCACCACUUUUGGCCACUUAACAUUCACGGUGGCUGGUUAAACAGGCGCUGAAGAUUAGGCCAUUCUCCCCGUGCUCCCAUU ......(((((((((((((((((.(((((....)).(((((((((....(.(((.......))).)....)))))))------))...))).))))))))))))))(((((...(((..((((..(((.(((......))).)))))))..))))))))..........)))...... ( -50.30, z-score = -2.02, R) >droYak2.chr3R 12503436 158 + 28832112 AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCUAAAGAAACGCCACCCAUCACCCACCGAUCGCCCCU-------------------UUUUCUGCACCACUUU-AACCACUUAACAUUCAUGGUUGCUGGUUAAGCGGGCUCUGAAGAUUAGUCCAUUUUCCCCGCGCUCCAAUU ......(((((((((((((........((....))...(((((........(((.......))).......-------------------.)))))))))))))))-(((((...........))))))))((....(((((....((((((......)))))))))))...)).... ( -42.29, z-score = -2.24, R) >droEre2.scaffold_4770 4316800 157 - 17746568 AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCCAAAGAAACGCCACCCAUCACCCACCAAUCGCCCCU--------------------UUUUUGCACCACUUU-GGCCACUUAACAUUCAUGGUUGCUGGUUAAGCAGGCGCCGAAGAUUAGUCCAUUUUCCCCGUGCUCCCAUU ........(((((((((((((((....((....)).........((...................))....--------------------.))))))))))))))-)..............((((.((((.....))))(((((.((((((......))))))...))))).)))). ( -39.21, z-score = -0.84, R) >droAna3.scaffold_13340 19485939 148 - 23697760 AUUCAAACCAAAGUGGUGCAAAAAAAAAGAUGAGC--AAGACCCACCAACAAUGCCUCCCCACCCACCCGACCAGCU--------CCCUUUUCCGUGCAUCGCUUUUGCCCACUUAACAUUCAAGAUUGCAUGUUAAGUUGGCCCAAACGCAGCGGCC-------------------- .......(((((((((((((....(((((..((((--.....................................)))--------).)))))...))))))))))).(((.(((((((((.((....)).))))))))).)))...........))..-------------------- ( -35.79, z-score = -2.54, R) >consensus AUUCAAAGCGAAGUGGUGCAAAAUGAUGCGCAAGCCAAAGAAACGCCACCCAUCACCCACCGAUCGCCCCU___U_U____________UUUUUUUGCACCACUUU_GGCCACUUAACAUUCACGGUUGCUGGUUAAACAGGCGCUGAAGAUUAGGCCAUUCUCCCCGUGCUCCCAUU ......(((((((((((((((((....((....))....(......).............................................)))))))))))))).((((......(......)....(((......))).............))))...........)))...... (-21.94 = -23.63 + 1.70)
| Location | 8,185,190 – 8,185,297 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Shannon entropy | 0.36150 |
| G+C content | 0.33180 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -15.00 |
| Energy contribution | -14.79 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
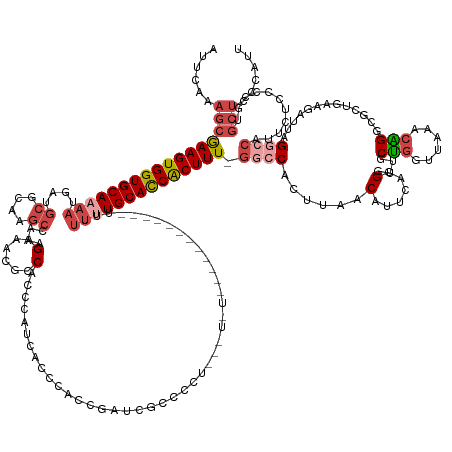
>dm3.chr3R 8185190 107 + 27905053 UAUAAAUGGGUUACAGAUUGCUCUUAAAAUAUAUACAUACAUAGGUGCAAAUUCACGAGUAUCUACUUAAAUCAAAAAUUCUAGCAA-----------GUCGCUAGAAUUCUACUAAU ......(((((...((((.((((........(((......))).(((......)))))))))))))))).......(((((((((..-----------...)))))))))........ ( -17.90, z-score = -1.74, R) >droSec1.super_0 7358201 94 - 21120651 UAUAAAUGGGCUACAGAUUGCUCCUAAAAUGUGUAUGUACUUAGGUGCAAAUUCUCGAGUACGUACUCAAAUCAAAAA--------------------GCAGCCGCAAUUAAGA---- ..(((.((((((.......(((.......((.(((((((((..((........))..))))))))).))........)--------------------)))))).)).)))...---- ( -22.07, z-score = -1.85, R) >droSim1.chr3R 14313674 114 - 27517382 UAUAAAUGGGCUACAGAUUGCUCCUAAAAUAUGUAUUUACUUAGGUGCAAAUUCUCGAGUACGUACUCAAAUCAUAAAUUCUAGUAAGUCACUUCAAAGCAGCCGCAAUUAAAA---- ..(((.((((((.(...((((.(((((.............))))).))))......((((((.((((..(((.....)))..)))).)).))))....).)))).)).)))...---- ( -20.62, z-score = -1.32, R) >consensus UAUAAAUGGGCUACAGAUUGCUCCUAAAAUAUGUAUAUACUUAGGUGCAAAUUCUCGAGUACGUACUCAAAUCAAAAAUUCUAG_AA___________GCAGCCGCAAUUAAAA____ ........((((.(...((((.(((((.............))))).))))......((((....))))..............................).)))).............. (-15.00 = -14.79 + -0.21)

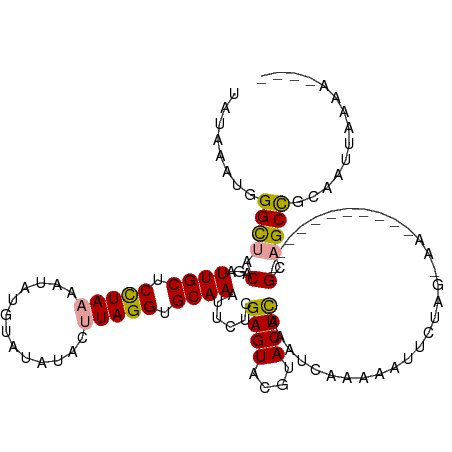
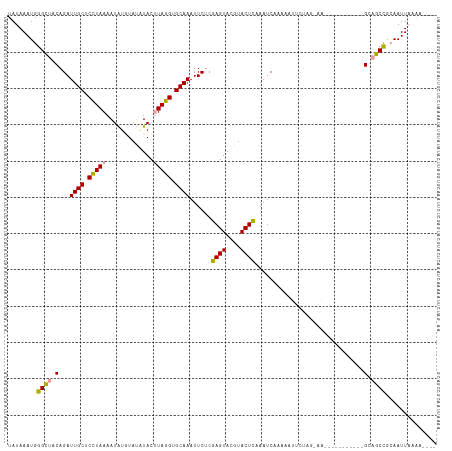
| Location | 8,185,190 – 8,185,297 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Shannon entropy | 0.36150 |
| G+C content | 0.33180 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -13.49 |
| Energy contribution | -15.60 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8185190 107 - 27905053 AUUAGUAGAAUUCUAGCGAC-----------UUGCUAGAAUUUUUGAUUUAAGUAGAUACUCGUGAAUUUGCACCUAUGUAUGUAUAUAUUUUAAGAGCAAUCUGUAACCCAUUUAUA ......(((((((((((...-----------..)))))))))))(((..((.(((.((((..(((......)))....)))).))).))..)))(((....))).............. ( -20.80, z-score = -1.51, R) >droSec1.super_0 7358201 94 + 21120651 ----UCUUAAUUGCGGCUGC--------------------UUUUUGAUUUGAGUACGUACUCGAGAAUUUGCACCUAAGUACAUACACAUUUUAGGAGCAAUCUGUAGCCCAUUUAUA ----...(((.((.((((((--------------------.......(((((((....)))))))...((((.((((((...........)))))).))))...)))))))).))).. ( -26.30, z-score = -3.45, R) >droSim1.chr3R 14313674 114 + 27517382 ----UUUUAAUUGCGGCUGCUUUGAAGUGACUUACUAGAAUUUAUGAUUUGAGUACGUACUCGAGAAUUUGCACCUAAGUAAAUACAUAUUUUAGGAGCAAUCUGUAGCCCAUUUAUA ----...(((.((.((((((...((..((.(((.((((((..((((.(((((((....))))))).((((((......)))))).))))))))))))))).)).)))))))).))).. ( -28.40, z-score = -2.76, R) >consensus ____UUUUAAUUGCGGCUGC___________UU_CUAGAAUUUUUGAUUUGAGUACGUACUCGAGAAUUUGCACCUAAGUAAAUACAUAUUUUAGGAGCAAUCUGUAGCCCAUUUAUA ..............((((((...........................(((((((....)))))))...((((.((((((...........)))))).))))...))))))........ (-13.49 = -15.60 + 2.11)

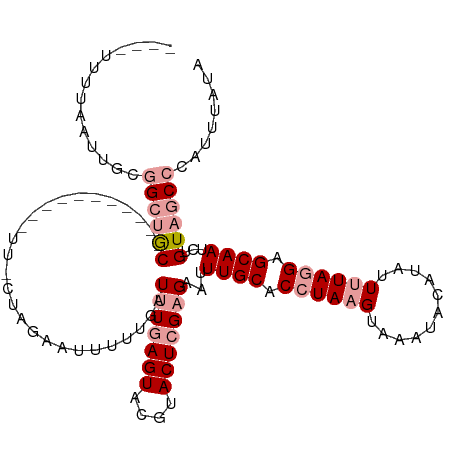
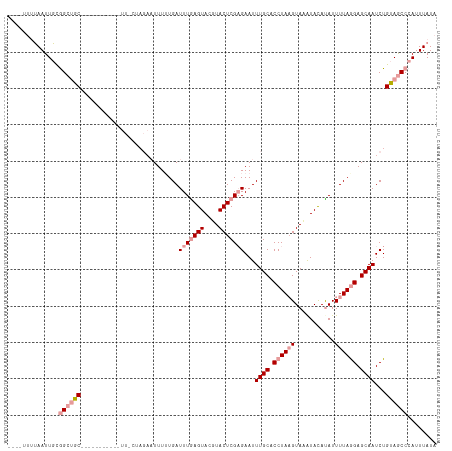
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:05 2011