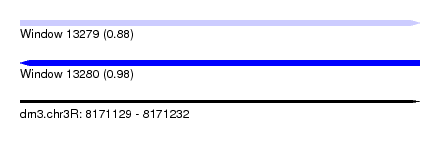
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,171,129 – 8,171,232 |
| Length | 103 |
| Max. P | 0.975506 |

| Location | 8,171,129 – 8,171,232 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.38860 |
| G+C content | 0.27852 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.55 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
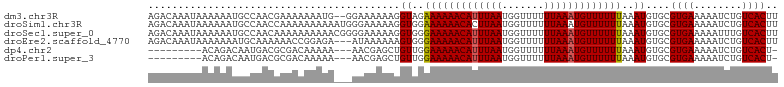
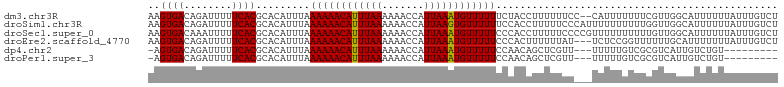
>dm3.chr3R 8171129 103 + 27905053 AGACAAAUAAAAAAUGCCAACGAAAAAAAUG--GGAAAAAAGGUAGAAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACUU .((((........((((((.(.(......).--)............(((((((((((((.......)))))))))))))...)).))))........)))).... ( -16.49, z-score = -1.22, R) >droSim1.chr3R 14299751 105 - 27517382 AGACAAAUAAAAAAUGCCAACCAAAAAAAAAAUGGGAAAAAGGUGGAAAAAACACUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACUU .((((........((((((.(((.........)))((((((.((..(((((((.(.....))))))))..)).))))))...)).))))........)))).... ( -19.39, z-score = -1.77, R) >droSec1.super_0 7344208 105 - 21120651 AGACAAAUAAAAAAUGCCAACAAAAAAAAAACGGGGAAAAAGGUGGGAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUUUGUCACUU .(((((((.....((((((...........((..........))..(((((((((((((.......)))))))))))))...)).)))).....))))))).... ( -20.40, z-score = -1.98, R) >droEre2.scaffold_4770 4302892 102 - 17746568 AGACAAAUAAAAAAAUGCAAAAAACCGGAGA---AUAAAAAAGUGGGAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACUU .((((.........(((((.....(((....---.........)))(((((((((((((.......))))))))))))).....)))))........)))).... ( -19.05, z-score = -2.19, R) >dp4.chr2 22967320 92 + 30794189 ---------ACAGACAAUGACGCGACAAAAA---AACGAGCUGUUGGAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACU- ---------(((((...(.((((.(((....---(((.....))).(((((((((((((.......)))))))))))))...))))))).)....)))))....- ( -20.50, z-score = -1.79, R) >droPer1.super_3 5744209 92 + 7375914 ---------ACAGACAAUGACGCGACAAAAA---AACGAGCUGUUGGAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACU- ---------(((((...(.((((.(((....---(((.....))).(((((((((((((.......)))))))))))))...))))))).)....)))))....- ( -20.50, z-score = -1.79, R) >consensus AGACAAAUAAAAAAUGCCAACGAAAAAAAAA___AAAAAAAGGUGGGAAAAACAUUUAAUGGUUUUUUAAAUGUUUUUUAAAUGUGCGUGAAAAAUCUGUCACUU ..........................................((..(((((((((((((.......)))))))))))))..))....((((........)))).. (-12.60 = -12.55 + -0.05)
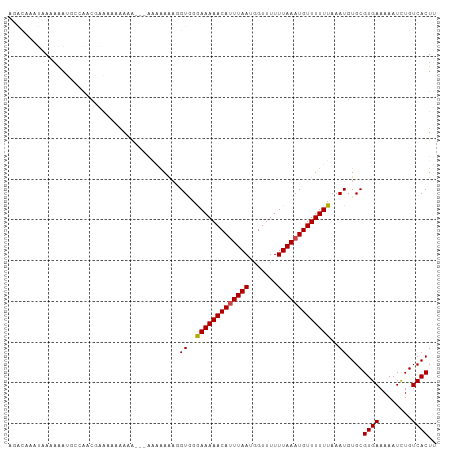
| Location | 8,171,129 – 8,171,232 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.45 |
| Shannon entropy | 0.38860 |
| G+C content | 0.27852 |
| Mean single sequence MFE | -16.42 |
| Consensus MFE | -10.64 |
| Energy contribution | -10.50 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
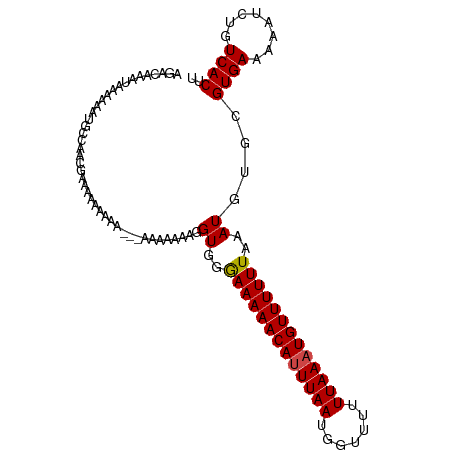
>dm3.chr3R 8171129 103 - 27905053 AAGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUUCUACCUUUUUUCC--CAUUUUUUUCGUUGGCAUUUUUUAUUUGUCU ....(((((((.......((......(((((((((((((.......))))))))))))).............--((..........)))).......))))))). ( -14.94, z-score = -2.35, R) >droSim1.chr3R 14299751 105 + 27517382 AAGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAGUGUUUUUUCCACCUUUUUCCCAUUUUUUUUUUGGUUGGCAUUUUUUAUUUGUCU ....(((((((.......((.((...(((((((((((((.......)))))))))))))...........(((.........))).)))).......))))))). ( -17.84, z-score = -2.36, R) >droSec1.super_0 7344208 105 + 21120651 AAGUGACAAAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUCCCACCUUUUUCCCCGUUUUUUUUUUGUUGGCAUUUUUUAUUUGUCU ....(((((((.......((.......((((((((((((.......))))))))))))..............((............)))).......))))))). ( -13.14, z-score = -1.28, R) >droEre2.scaffold_4770 4302892 102 + 17746568 AAGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUCCCACUUUUUUAU---UCUCCGGUUUUUUGCAUUUUUUUAUUUGUCU ....(((((((.......(((......((((((((((((.......)))))))))))).((..........---.....)).....)))........))))))). ( -14.62, z-score = -2.25, R) >dp4.chr2 22967320 92 - 30794189 -AGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUCCAACAGCUCGUU---UUUUUGUCGCGUCAUUGUCUGU--------- -....((((((.....(((((((....((((((((((((.......))))))))))))..(((.....)))---....))).))))....))))))--------- ( -19.00, z-score = -2.71, R) >droPer1.super_3 5744209 92 - 7375914 -AGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUCCAACAGCUCGUU---UUUUUGUCGCGUCAUUGUCUGU--------- -....((((((.....(((((((....((((((((((((.......))))))))))))..(((.....)))---....))).))))....))))))--------- ( -19.00, z-score = -2.71, R) >consensus AAGUGACAGAUUUUUCACGCACAUUUAAAAAACAUUUAAAAAACCAUUAAAUGUUUUUCCCACCUUUUUUC___UUUUUGUUUCGUUGGCAUUUUUUAUUUGUCU ..((((........)))).........((((((((((((.......))))))))))))............................................... (-10.64 = -10.50 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:02 2011