
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,168,526 – 7,168,619 |
| Length | 93 |
| Max. P | 0.619476 |

| Location | 7,168,526 – 7,168,619 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.41 |
| Shannon entropy | 0.53086 |
| G+C content | 0.52769 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -15.25 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7168526 93 - 23011544 AAACCGGAACAGAUCCAGCACACUAAA---UUCCUCUUGGCAGAUAAAAGUUGUCGGCCAUUUCACGGCGCAGUGAGCUCGGUGACGUGCCACGCC----- ...(((((.....)))...........---.......((((.((((.....)))).))))......)).((.(((.((.((....)).))))))).----- ( -25.00, z-score = -0.20, R) >droSim1.chr2L 6958288 93 - 22036055 AAACCGGAACAGAUCUAGCGCACUAAA---UUCCUCUUGGCAGAUAAAAGUUGUCGGCCAUUUCACGGCGCAGUGAGCUCGGUGACGUGCCACGCC----- .....((..........((((......---.......((((.((((.....)))).)))).......)))).(((.((.((....)).))))).))----- ( -25.75, z-score = -0.07, R) >droSec1.super_3 2692069 76 - 7220098 -----------------GCACACUAAA---UUCCUCUUGGCAGAUAAAAGUUGUCGGCCAUUUCACGGCGCAGUGAGCUCGGUGACGUGCCACGCC----- -----------------((((......---.......((((.((((.....)))).))))..((((.(.((.....)).).)))).))))......----- ( -21.70, z-score = -0.34, R) >droYak2.chr2L 16588198 93 + 22324452 AAACCGGAACAGAUCUCGCACACCAAA---UUCCACUUGGCAGAUAAAACUUGUCGGCCAUUUCACGGCGCAGUGAGCUCGGUGACGUGCCACGCC----- ...((((((..(....)..........---.......((((.((((.....)))).)))).))).))).((.(((.((.((....)).))))))).----- ( -24.40, z-score = -0.14, R) >droEre2.scaffold_4929 16083754 93 - 26641161 AAACCGGAACAGAUCCAACAAACCAAA---UUUCCCUUGGCAGAUAAAACUUGUCGGCCAUUUCACGGCUCUGUGAGCUCAGUGACGUGCCACGCC----- .....(((.....)))...........---........(((.((((.....))))(((.((.(((((((((...)))))..)))).)))))..)))----- ( -22.30, z-score = -0.75, R) >droAna3.scaffold_12943 1018805 93 + 5039921 AAGCCGGAACGGGUCCGACAAACUAAA---AUCCGCUUGGCCGACGAAAGUUGUCGGCCAUUUCACGGGACAAUGAGCUGGGUGACAGGCCACGCC----- ..(.((((.....)))).)........---....((.((((((((....)))((((.(((.((((........)))).))).)))).))))).)).----- ( -32.70, z-score = -1.11, R) >dp4.chr4_group2 808310 96 - 1235136 CAUCAAGAAAGAAUC--ACAAACUAAA---UUCCGUUUGGCAGACGAAAGUUGUCGGCCAUUUUACGGCACAGUGAGCUCUGGGAGGAGCCACGCCACGCC ...............--..........---....((.((((.(((....)))((((.........))))...(((.(((((....)))))))))))).)). ( -26.30, z-score = -1.06, R) >droPer1.super_8 872849 85 + 3966273 -----------AAUC--ACAAACUAAA---UUCCGUUUGGCAGACGAAAGUUGUCGGCCAUUUUACGGCACAGUGAGCUCUGGGAGGAGUCACGCCACGCC -----------....--..........---....((.((((.(((....)))((((.........))))...((((.((((....)))))))))))).)). ( -24.20, z-score = -0.90, R) >droWil1.scaffold_180772 3256869 85 + 8906247 ------GAAGAAAAAUGGCAAACUAAAUUUUUCCGUCAGGCAGACGAAAGUCGUCGGCCAUUUCACGCCACAGUGAACACUUUGCCCUAAC---------- ------..........((((((................(((.((((.....)))).)))..(((((......)))))...)))))).....---------- ( -23.20, z-score = -2.62, R) >consensus AAACCGGAACAGAUC__GCAAACUAAA___UUCCGCUUGGCAGAUAAAAGUUGUCGGCCAUUUCACGGCACAGUGAGCUCGGUGACGUGCCACGCC_____ .....................................((((.(((((...))))).))))......(((...(((.((.((....)).))))))))..... (-15.25 = -15.88 + 0.63)

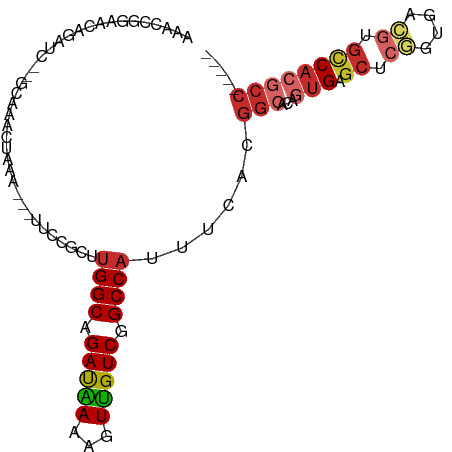

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:04 2011