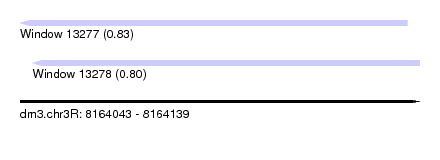
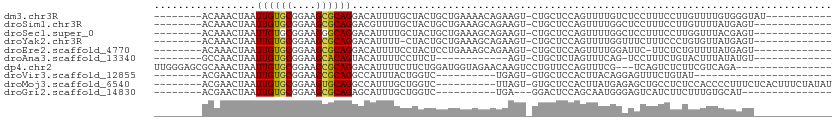
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,164,043 – 8,164,139 |
| Length | 96 |
| Max. P | 0.826702 |

| Location | 8,164,043 – 8,164,136 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.92 |
| Shannon entropy | 0.70691 |
| G+C content | 0.45250 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.48 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8164043 93 - 27905053 --------ACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUGCUACUGCUGAAAACAGAAGU-CUGCUCCAGUUUUGUCUCCUUUCCUUGUUUUGUGGGUAU----------- --------(((((.(((.(((((....)))))(((((....((.(((.(((....))).)))-..))........))))).......))).)))))......----------- ( -20.50, z-score = 0.31, R) >droSim1.chr3R 14292631 91 + 27517382 --------ACAAACUAAUUGUGCGGAAGCGCAGGACGUUUUGCUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGCUCCUUUCCUUGUUUUAUGAGU------------- --------((((......(((((....)))))(((......((((..((((..(((((....-))))).))))..)))).....))))))).........------------- ( -28.80, z-score = -1.80, R) >droSec1.super_0 7337463 91 + 21120651 --------ACAAACUAAUUGUGCGGAAGGGCAGGACAUUUUGCUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGCUCCUUUCCUUGGUUUACGAGU------------- --------..(((((((.(((.(....).)))(((......((((..((((..(((((....-))))).))))..)))).....))))))))))......------------- ( -28.50, z-score = -1.29, R) >droYak2.chr3R 12482435 90 - 28832112 --------ACAAACUAAUUGUGCGGAAGCGCAGGACAUUUU-CUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGUUUCUUUCCCUGUGUUAUGAGU------------- --------.(((((....(((((....)))))(((......-(((..((((..(((((....-))))).))))..)))......)))....))).))...------------- ( -25.30, z-score = -1.28, R) >droEre2.scaffold_4770 4296191 90 + 17746568 --------ACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCCUACUCCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGAUUC-UUCUCUGUUUUAUGAGU------------- --------...........((((....))))((((.....))))((((...((((((((.(.-....(((......)))...-..)))))))))..))))------------- ( -25.40, z-score = -1.75, R) >droAna3.scaffold_13340 19465357 78 + 23697760 --------GCCAACUAAUUGUGCGGAAGCACAGUACAUUUUCCUUCU------------AGU-CUGCUCUAGUUUCAG-UCCUUUCUGUACUUUAUAUGU------------- --------((..(((((((((((....)))))))............)------------)))-..))...(((..(((-......))).)))........------------- ( -16.30, z-score = -1.96, R) >dp4.chr2 22957629 94 - 30794189 UUGGGAGCGCAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCUUCUGGAUGGUAGAACAAGUCCUGUUCCAGUUUCG---UCAGUCUCUUCGUCAGA---------------- ..((((((((((.....)))))).((((((((((((...((((.((....)).))))...)))))))....))))).---....)))).........---------------- ( -27.40, z-score = -0.89, R) >droVir3.scaffold_12855 9684765 71 - 10161210 --------ACGAACUAAUUGUGCGGAAGCGCAGGCCAUUUACUGGUC----------UGAGU-GUGCUCCACUUACAGGAGUUUCUGUAU----------------------- --------...........((((((((((.(((((((.....)))))----------)).))-..(((((.......)))))))))))))----------------------- ( -24.40, z-score = -2.34, R) >droMoj3.scaffold_6540 24719603 94 - 34148556 --------ACGAACUAAUUGUGCGGAAGUGCAGGCCAUUUGCUGGUC----------UUAGU-GUGCUCCACUUAUGAGAGCUGCCUCUCCACCCCUUUCUCACUUUCUAUAU --------....(((((.((..(....)..))(((((.....)))))----------)))))-(((..........(((((....)))))...........)))......... ( -21.00, z-score = -0.23, R) >droGri2.scaffold_14830 2065236 77 - 6267026 --------ACGAACUAAUUGUGCGGAAGCGCAGAGCAUUUGCUGGUC----------UGA---GGACUCCAGCAAUGGGAGUCAUCUUCUUUGUGCAU--------------- --------.........((((((....)))))).(((((((((((((----------...---.))..))))))).(((((....)))))..))))..--------------- ( -26.40, z-score = -1.66, R) >consensus ________ACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCCUACUGCUGAAAGCAGAAGU_CUGCUCCAGUUUUGGCUCCUUUCUUUGUUUUAUGAGU_____________ .................((((((....))))))................................................................................ ( -9.55 = -9.48 + -0.07)


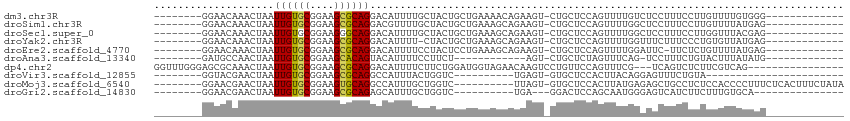
| Location | 8,164,046 – 8,164,139 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.55 |
| Shannon entropy | 0.69674 |
| G+C content | 0.46462 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -9.55 |
| Energy contribution | -9.48 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8164046 93 - 27905053 --------GGAACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUGCUACUGCUGAAAACAGAAGU-CUGCUCCAGUUUUGUCUCCUUUCCUUGUUUUGUGGG------------- --------((((((((((...(((((....)))))(((......((.(((.(((....))).)))-..))))))).))))).)))...(((........)))------------- ( -23.60, z-score = -0.37, R) >droSim1.chr3R 14292632 93 + 27517382 --------GGAACAAACUAAUUGUGCGGAAGCGCAGGACGUUUUGCUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGCUCCUUUCCUUGUUUUAUGAG------------- --------(((((((......(((((....)))))(((......((((..((((..(((((....-))))).))))..)))).....)))))))))).....------------- ( -32.10, z-score = -2.63, R) >droSec1.super_0 7337464 93 + 21120651 --------GGAACAAACUAAUUGUGCGGAAGGGCAGGACAUUUUGCUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGCUCCUUUCCUUGGUUUACGAG------------- --------.....(((((((.(((.(....).)))(((......((((..((((..(((((....-))))).))))..)))).....)))))))))).....------------- ( -28.50, z-score = -1.16, R) >droYak2.chr3R 12482436 92 - 28832112 --------GGAACAAACUAAUUGUGCGGAAGCGCAGGACAUUUU-CUACUGCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGUUUCUUUCCCUGUGUUAUGAG------------- --------((((.(((((((((((((....))))))........-.....((((..(((((....-))))).)))).)))))))..))))............------------- ( -27.20, z-score = -1.59, R) >droEre2.scaffold_4770 4296192 92 + 17746568 --------GGAACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCCUACUCCUGAAAGCAGAAGU-CUGCUCCAGUUUUGGAUUC-UUCUCUGUUUUAUGAG------------- --------((((........((((((....))))))......))))..(((...((((((((.(.-....(((......)))...-..)))))))))..)))------------- ( -25.74, z-score = -1.51, R) >droAna3.scaffold_13340 19465358 80 + 23697760 --------GAUGCCAACUAAUUGUGCGGAAGCACAGUACAUUUUCCUUCU------------AGU-CUGCUCUAGUUUCAG-UCCUUUCUGUACUUUAUAUG------------- --------((.((..(((((((((((....)))))))............)------------)))-..)))).(((..(((-......))).))).......------------- ( -17.00, z-score = -1.95, R) >dp4.chr2 22957630 96 - 30794189 GGUUUGGGAGCGCAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCUUCUGGAUGGUAGAACAAGUCCUGUUCCAGUUUCG---UCAGUCUCUUCGUCAG---------------- .....((((((((((.....)))))).((((((((((((...((((.((....)).))))...)))))))....))))).---....))))........---------------- ( -27.40, z-score = -0.91, R) >droVir3.scaffold_12855 9684766 73 - 10161210 --------GGUACGAACUAAUUGUGCGGAAGCGCAGGCCAUUUACUGGUC----------UGAGU-GUGCUCCACUUACAGGAGUUUCUGUA----------------------- --------.((((((.....))))))(((.((((((((((.....)))))----------....)-)))))))...((((((....))))))----------------------- ( -23.80, z-score = -1.50, R) >droMoj3.scaffold_6540 24719604 96 - 34148556 --------GGAACGAACUAAUUGUGCGGAAGUGCAGGCCAUUUGCUGGUC----------UUAGU-GUGCUCCACUUAUGAGAGCUGCCUCUCCACCCCUUUCUCACUUUCUAUA --------(((.((.(((((.((..(....)..))(((((.....)))))----------)))))-.)).)))......(((((....)))))...................... ( -25.60, z-score = -1.03, R) >droGri2.scaffold_14830 2065237 79 - 6267026 --------GGAACGAACUAAUUGUGCGGAAGCGCAGAGCAUUUGCUGGUC----------UGA---GGACUCCAGCAAUGGGAGUCAUCUUCUUUGUGCA--------------- --------............((((((....)))))).(((((((((((((----------...---.))..))))))).(((((....)))))..)))).--------------- ( -26.40, z-score = -1.30, R) >consensus ________GGAACAAACUAAUUGUGCGGAAGCGCAGGACAUUUUCCUACUGCUGAAAGCAGAAGU_CUGCUCCAGUUUUGGCUCCUUUCUUUGUUUUAUGAG_____________ ....................((((((....))))))............................................................................... ( -9.55 = -9.48 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:09:00 2011