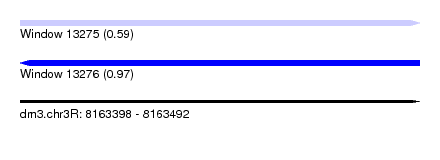
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,163,398 – 8,163,492 |
| Length | 94 |
| Max. P | 0.971047 |

| Location | 8,163,398 – 8,163,492 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.38 |
| Shannon entropy | 0.52384 |
| G+C content | 0.41907 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -10.25 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8163398 94 + 27905053 ----------CGCUAGUUGAGGUCACAACUGGCA-AUUAG------UUUUAUGGUGGGUGAAGCGACCUCAAAGAAAAUUGAGGGCU--AUGAGGUGGCAGGUUAUUAAAUCC ----------.((((((((......)))))))).-....(------((((((.....)))))))..((((((......))))))(((--((...))))).............. ( -26.70, z-score = -2.09, R) >droSim1.chr3R 14291961 94 - 27517382 ----------CGCUAGUUGAGGUCACAACUGGCA-AUUAG------UUUUAUGGUGGGUGAAGCGACCUCAAGGAAAAUUGUGGGCU--AUGAGGUGGCAGGUUAUUAAAUCA ----------.((((((((......)))))))).-.....------.....((((..((((.((.((((((.((...........))--.)))))).))...))))...)))) ( -26.70, z-score = -1.98, R) >droSec1.super_0 7336801 94 - 21120651 ----------CGCUAGUUGAGGUCACAACUGGCA-AUUAG------UUUUAUGGUGGGUGAAGCGACCUCAAGGAAAAUUGUGGGCU--AUGAGGUGGCAGGUUAUUAAAUCA ----------.((((((((......)))))))).-.....------.....((((..((((.((.((((((.((...........))--.)))))).))...))))...)))) ( -26.70, z-score = -1.98, R) >droYak2.chr3R 12481752 94 + 28832112 ----------CGCUAGUUGAGGUCACAACUGGCA-AUUAG------UUUUAUGGUGGGUGAAGCGACCUCAAGGAAAGUUGAGGGCU--AUAAGGUGGCAGGUUAUUAAAUCA ----------.((((((((......)))))))).-....(------((((((.....)))))))..((((((......))))))(((--((...))))).((((....)))). ( -27.20, z-score = -2.39, R) >droEre2.scaffold_4770 4295510 94 - 17746568 ----------CGCUAGUUGAGGUCACAACUGGCA-AUUAG------UUUUAUGGUGGGUGAAGCGACCUCAAGGAAAAUUGAGGGCU--AUGUGGUGGCAGGUUAUUAAAUCA ----------.((((((((......)))))))).-....(------((((((.....)))))))..((((((......))))))(((--((...))))).((((....)))). ( -28.10, z-score = -2.48, R) >dp4.chr2 22956841 95 + 30794189 ----------CGAUAGUUACGGUCAUGCCUAGAA-AUUAC------UUUAAUGGUGGGUGAAGCGACCUUACAAGGAUUUUGGGAUUG-AGGUCAAGGGAGAUCAGAGAAUCA ----------..........((((.(.(((....-.((((------((.......))))))...(((((((((((...))))....))-))))).))).)))))......... ( -18.10, z-score = 0.65, R) >droGri2.scaffold_14830 2064489 112 + 6267026 CAACAGAAAAGACUAAAUGUACAUAUCUAUGGAAUAUUAAAUACUCUUUUCUGACAGAUUUCA-GAUCACACAGAUAGUUCCGACUUCAAUUAAAUUUUGUGUAAUUUAAUUU ...(((((((((.(((((((.(((....)))..)))))...)).))))))))).........(-(((.((((((((((((........)))))...))))))).))))..... ( -18.30, z-score = -1.07, R) >consensus __________CGCUAGUUGAGGUCACAACUGGCA_AUUAG______UUUUAUGGUGGGUGAAGCGACCUCAAGGAAAAUUGAGGGCU__AUGAGGUGGCAGGUUAUUAAAUCA ...........((((((((......))))))))...................(((.(......).)))............................................. (-10.25 = -10.40 + 0.15)
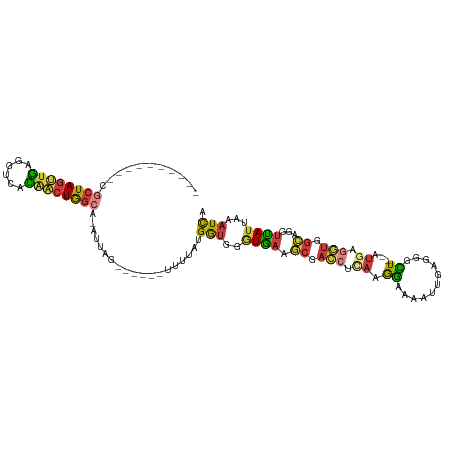

| Location | 8,163,398 – 8,163,492 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.38 |
| Shannon entropy | 0.52384 |
| G+C content | 0.41907 |
| Mean single sequence MFE | -15.85 |
| Consensus MFE | -9.87 |
| Energy contribution | -9.89 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8163398 94 - 27905053 GGAUUUAAUAACCUGCCACCUCAU--AGCCCUCAAUUUUCUUUGAGGUCGCUUCACCCACCAUAAAA------CUAAU-UGCCAGUUGUGACCUCAACUAGCG---------- ..............((.((((((.--((...........)).)))))).))................------.....-.((.(((((......))))).)).---------- ( -17.50, z-score = -2.16, R) >droSim1.chr3R 14291961 94 + 27517382 UGAUUUAAUAACCUGCCACCUCAU--AGCCCACAAUUUUCCUUGAGGUCGCUUCACCCACCAUAAAA------CUAAU-UGCCAGUUGUGACCUCAACUAGCG---------- ..............((.((((((.--................)))))).))................------.....-.((.(((((......))))).)).---------- ( -17.03, z-score = -2.63, R) >droSec1.super_0 7336801 94 + 21120651 UGAUUUAAUAACCUGCCACCUCAU--AGCCCACAAUUUUCCUUGAGGUCGCUUCACCCACCAUAAAA------CUAAU-UGCCAGUUGUGACCUCAACUAGCG---------- ..............((.((((((.--................)))))).))................------.....-.((.(((((......))))).)).---------- ( -17.03, z-score = -2.63, R) >droYak2.chr3R 12481752 94 - 28832112 UGAUUUAAUAACCUGCCACCUUAU--AGCCCUCAACUUUCCUUGAGGUCGCUUCACCCACCAUAAAA------CUAAU-UGCCAGUUGUGACCUCAACUAGCG---------- ........................--(((((((((......))))))..)))...............------.....-.((.(((((......))))).)).---------- ( -15.50, z-score = -2.10, R) >droEre2.scaffold_4770 4295510 94 + 17746568 UGAUUUAAUAACCUGCCACCACAU--AGCCCUCAAUUUUCCUUGAGGUCGCUUCACCCACCAUAAAA------CUAAU-UGCCAGUUGUGACCUCAACUAGCG---------- ........................--(((((((((......))))))..)))...............------.....-.((.(((((......))))).)).---------- ( -15.60, z-score = -2.05, R) >dp4.chr2 22956841 95 - 30794189 UGAUUCUCUGAUCUCCCUUGACCU-CAAUCCCAAAAUCCUUGUAAGGUCGCUUCACCCACCAUUAAA------GUAAU-UUCUAGGCAUGACCGUAACUAUCG---------- .(((....((.((..(((.(((((-(((...........)))..)))))((((............))------))...-....)))...)).)).....))).---------- ( -7.90, z-score = 1.32, R) >droGri2.scaffold_14830 2064489 112 - 6267026 AAAUUAAAUUACACAAAAUUUAAUUGAAGUCGGAACUAUCUGUGUGAUC-UGAAAUCUGUCAGAAAAGAGUAUUUAAUAUUCCAUAGAUAUGUACAUUUAGUCUUUUCUGUUG .(((((((((......)))))))))((..(((((((.......))..))-)))..))...(((((((((.((....(((((.....))))).......)).)))))))))... ( -20.40, z-score = -1.16, R) >consensus UGAUUUAAUAACCUGCCACCUCAU__AGCCCUCAAUUUUCCUUGAGGUCGCUUCACCCACCAUAAAA______CUAAU_UGCCAGUUGUGACCUCAACUAGCG__________ .........................................((((((((((....................................))))))))))................ ( -9.87 = -9.89 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:59 2011