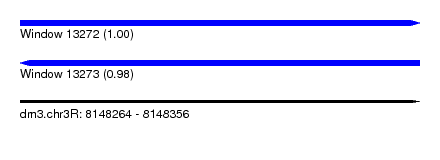
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,148,264 – 8,148,356 |
| Length | 92 |
| Max. P | 0.998017 |

| Location | 8,148,264 – 8,148,356 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.37085 |
| G+C content | 0.48999 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
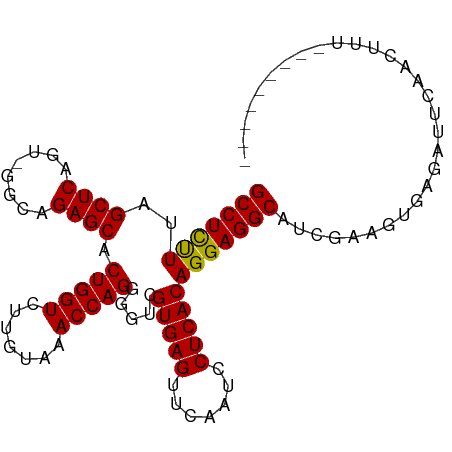
>dm3.chr3R 8148264 92 + 27905053 ---------AAAGUUGAAUCUCAAUUCGUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC ---------..(((((.....)))))....(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -27.80, z-score = -2.13, R) >droSim1.chr3R 14279721 92 - 27517382 ---------AAAGUUGAAUCUCAAUUCAAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC ---------...(((((((....)))))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -30.30, z-score = -3.60, R) >droSec1.super_0 7321924 92 - 21120651 ---------AAAGUUGAAUCUCAAUUCGAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC ---------...(((((((....)))))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -30.00, z-score = -3.38, R) >droYak2.chr3R 12466160 91 + 28832112 ---------AAAGUUGAAUCUCC-UUCGAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC ---------...((((((.....-))))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -29.00, z-score = -2.70, R) >droEre2.scaffold_4770 4279403 91 - 17746568 ---------AAAGUUGAAUCCCA-UUCGAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC ---------...(((((((...)-))))))(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -29.20, z-score = -3.05, R) >droAna3.scaffold_13340 19439019 86 - 23697760 ---------------AAAGGAAACACUCUUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUACC-ACUGAGCUAAAGAGGC ---------------...(....)......(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -28.10, z-score = -2.86, R) >droPer1.super_3 5715267 92 + 7375914 --------AAAAAUAUAUGAA-ACUUCUAUGCCUCCUGUGAGAAUUGAACUCACGGCCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAGGAGGC --------.............-........((((((((((((.......))))).....((((((.....)))))).((((....-...))))..))))))) ( -29.90, z-score = -3.68, R) >dp4.chr2 22938217 92 + 30794189 --------AAAAAUAUAUGAA-ACUUCUAUGCCUCCUGUGAGAAUUGAACUCACGGCCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAGGAGGC --------.............-........((((((((((((.......))))).....((((((.....)))))).((((....-...))))..))))))) ( -29.90, z-score = -3.68, R) >droWil1.scaffold_181089 12189120 91 - 12369635 ----------GCAUCCCCCUUUGAACAUUUGCCUCCUGUGAGAAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAGAAGGC ----------....................((((.(((((((.......))))).....((((((.....)))))).((((....-...))))..)).)))) ( -23.10, z-score = -1.92, R) >droVir3.scaffold_12855 9654279 96 + 10161210 -----CAUUAAAAUAUCUUUUUACUUGACUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC -----.........................(((((.((((((.......))))))....((((((.....)))))).((((....-...))))....))))) ( -26.40, z-score = -2.63, R) >droMoj3.scaffold_6540 24699127 101 + 34148556 UAUUAACCAAAAAUAUCGAUUCAGUUCGUUGCCUCCUGUGAGAAUUGAACUCACGGCCCCUGGUUUACAAGACCAGUGCUCUGCC-ACUGAGCUAAAGAGGC ................(((......)))..((((((((((((.......)))))))...((((((.....)))))).((((....-...))))....))))) ( -28.70, z-score = -2.59, R) >anoGam1.chr2L 32141846 93 + 48795086 ---------CACGCACGCUUUCUCACCACUGCCUCCUGUGAGAAUCGAACUCACGACCGCUGGUUUACAAGACCAGCGCUCUGCCUACUGAGCUAAAGAGGC ---------.....................(((((.((((((.......))))))...(((((((.....)))))))((((........))))....))))) ( -30.20, z-score = -2.84, R) >consensus _________AAAGUAGAAUCUCACUUCGAUGCCUCCUGUGAGGAUUGAACUCACGACCCCUGGUUUACAAGACCAGUGCUCUGCC_ACUGAGCUAAAGAGGC ..............................(((((.((((((.......))))))....((((((.....)))))).((((........))))....))))) (-25.72 = -25.80 + 0.08)

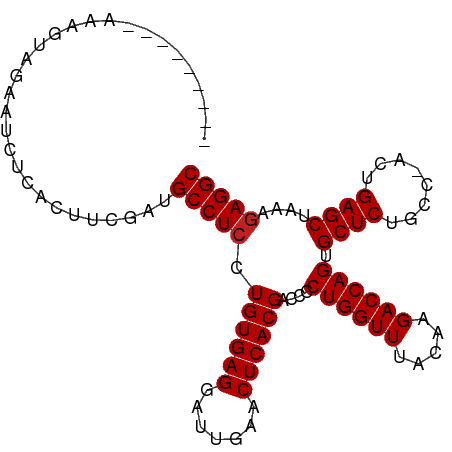
| Location | 8,148,264 – 8,148,356 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.87 |
| Shannon entropy | 0.37085 |
| G+C content | 0.48999 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.44 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8148264 92 - 27905053 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAACGAAUUGAGAUUCAACUUU--------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))...((((....))))......--------- ( -28.90, z-score = -1.37, R) >droSim1.chr3R 14279721 92 + 27517382 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUUGAAUUGAGAUUCAACUUU--------- (((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).((((((....))))))....--------- ( -30.20, z-score = -1.88, R) >droSec1.super_0 7321924 92 + 21120651 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUCGAAUUGAGAUUCAACUUU--------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))...((((....))))......--------- ( -28.90, z-score = -1.55, R) >droYak2.chr3R 12466160 91 - 28832112 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUCGAA-GGAGAUUCAACUUU--------- (((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).((...-.))...........--------- ( -29.10, z-score = -1.23, R) >droEre2.scaffold_4770 4279403 91 + 17746568 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUCGAA-UGGGAUUCAACUUU--------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))...(((-(...))))......--------- ( -27.80, z-score = -0.83, R) >droAna3.scaffold_13340 19439019 86 + 23697760 GCCUCUUUAGCUCAGU-GGUAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAAGAGUGUUUCCUUU--------------- ((((.....((((...-....)))).(((((.......))))))))).(((((.......)))))((((((((....))))))))..--------------- ( -28.10, z-score = -1.45, R) >droPer1.super_3 5715267 92 - 7375914 GCCUCCUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGCCGUGAGUUCAAUUCUCACAGGAGGCAUAGAAGU-UUCAUAUAUUUUU-------- (((((((..((((...-....))))...((((((........))))))(((((.......))))))))))))........-.............-------- ( -31.80, z-score = -2.98, R) >dp4.chr2 22938217 92 - 30794189 GCCUCCUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGCCGUGAGUUCAAUUCUCACAGGAGGCAUAGAAGU-UUCAUAUAUUUUU-------- (((((((..((((...-....))))...((((((........))))))(((((.......))))))))))))........-.............-------- ( -31.80, z-score = -2.98, R) >droWil1.scaffold_181089 12189120 91 + 12369635 GCCUUCUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUUCUCACAGGAGGCAAAUGUUCAAAGGGGGAUGC---------- (((((((..((((...-....)))).(((((.......))))).....(((((.......))))))))))))....((((......))))..---------- ( -28.50, z-score = -1.04, R) >droVir3.scaffold_12855 9654279 96 - 10161210 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAGUCAAGUAAAAAGAUAUUUUAAUG----- (((((((..((((...-....)))).(((((.......))))).....(((((.......)))))))))))).........................----- ( -27.60, z-score = -1.65, R) >droMoj3.scaffold_6540 24699127 101 - 34148556 GCCUCUUUAGCUCAGU-GGCAGAGCACUGGUCUUGUAAACCAGGGGCCGUGAGUUCAAUUCUCACAGGAGGCAACGAACUGAAUCGAUAUUUUUGGUUAAUA (((((((..((((...-....))))...((((((........))))))(((((.......))))))))))))....(((..((........))..))).... ( -32.70, z-score = -2.17, R) >anoGam1.chr2L 32141846 93 - 48795086 GCCUCUUUAGCUCAGUAGGCAGAGCGCUGGUCUUGUAAACCAGCGGUCGUGAGUUCGAUUCUCACAGGAGGCAGUGGUGAGAAAGCGUGCGUG--------- (((((((..(((.....))).((.(((((((.......))))))).))(((((.......)))))))))))).(((((......)).)))...--------- ( -34.30, z-score = -1.38, R) >consensus GCCUCUUUAGCUCAGU_GGCAGAGCACUGGUCUUGUAAACCAGGGGUCGUGAGUUCAAUCCUCACAGGAGGCAUCGAAGUGAGAUUCAACUUU_________ (((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).............................. (-28.70 = -28.44 + -0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:56 2011