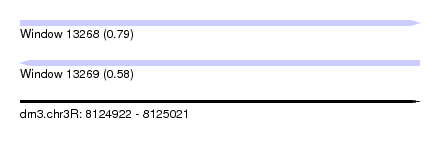
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,124,922 – 8,125,021 |
| Length | 99 |
| Max. P | 0.792454 |

| Location | 8,124,922 – 8,125,021 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Shannon entropy | 0.34383 |
| G+C content | 0.36372 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
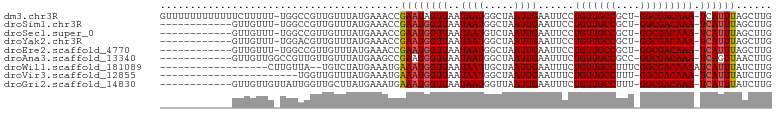
>dm3.chr3R 8124922 99 + 27905053 GUUUUUUUUUUUUCUUUUU-UGGCCGUUGUUUAUGAAACCGAAUAGUUAAUAAUGGCUAAUUGAAUUCCUGUUGCCGCU-GGCGACAAA-UCAUUUAGCUUG ...........(((....(-((((((((((((((........)))...))))))))))))..)))....(((((((...-)))))))..-............ ( -21.40, z-score = -1.87, R) >droSim1.chr3R 14255332 87 - 27517382 ------------GUUGUUU-UGGCCGUUGUUUAUGAAACCGAAUGGUUAAUAAUGGCUAAUUGAAUUCCUGUUGCCGCU-GGCGACAAA-UCAUUUAGCUUG ------------((((..(-(((((((((((.....((((....)))))))))))))))).........(((((((...-)))))))..-.....))))... ( -24.30, z-score = -2.47, R) >droSec1.super_0 7298536 87 - 21120651 ------------GUUGUUU-UGGCCGUUGUUUAUGAAACCGAAUGGUUAAUAAUGUCUAAUUGAAUUCCUGUUGCCGCU-GGCGACAAA-UCAUUUAGCUUG ------------......(-(((((((((((.....)))..)))))))))....(.((((.(((.....(((((((...-)))))))..-))).)))))... ( -18.00, z-score = -0.84, R) >droYak2.chr3R 12442346 87 + 28832112 ------------GUUGUUU-UGGACGUUGUUUAUGAAACCGAAUGGUUAAUAAUGGCUAAUUGAAUUCCUGUUGCCGCU-GGCGACAAA-UCAUUUAGCUUG ------------.......-....(((((((.....((((....)))))))))))(((((.(((.....(((((((...-)))))))..-))).)))))... ( -20.80, z-score = -1.52, R) >droEre2.scaffold_4770 4255501 87 - 17746568 ------------GUUGUUU-UGGCCGUUGUUUAUGAAACCGAAUGGUUAAUAAUGGCUAAUUGAAUUCCUGUUGCCGCU-GGCGACAAA-UCAUUUAGCUUG ------------((((..(-(((((((((((.....((((....)))))))))))))))).........(((((((...-)))))))..-.....))))... ( -24.30, z-score = -2.47, R) >droAna3.scaffold_13340 19416288 88 - 23697760 ------------GUUGUUGGCCGUUGUUGUUUAUGAAGCCGAAUGGUUAAUAAUGGCUAAUUGAAUUUCUGUUGCCGCC-GGCGACAAA-UCAGUUAACUUG ------------((((.((((((((((((.((((........)))).))))))))))))(((((.....(((((((...-)))))))..-)))))))))... ( -25.30, z-score = -2.22, R) >droWil1.scaffold_181089 207979 82 + 12369635 ------------------CUUGUUA--UGUCUAUGAAAUGAAAUGGUUAAUAAUUGCUAAUUGAAUUUCUGUUGCCUUUCGGCGACAAAAUCAUUUAUCUUG ------------------.......--........((((((..((((........))))..........(((((((....)))))))...))))))...... ( -15.30, z-score = -2.31, R) >droVir3.scaffold_12855 9613482 77 + 10161210 -----------------------UGGUUGUUUAUGAAAUGAAAUGGUUAAUAAUGGCUAAUUGAAUUUCUGUUGCCUUU-GGCGACAAA-UCAUUUAUCUUG -----------------------(((((......(((((..((((((((....))))).)))..)))))(((((((...-)))))))))-)))......... ( -16.10, z-score = -2.21, R) >droGri2.scaffold_14830 2022424 88 + 6267026 ------------GUUGUUGUUAUUGGUUGCUUAUGAAAUGAAAUGGUUAAUAAUGGUUAAUUGAAUUUCUGUUGCCUUU-GGCGACAAA-UCAUUUAUCUUG ------------.(((((((((..(((.((....(((((....((((((((....)))))))).))))).)).)))..)-)))))))).-............ ( -19.80, z-score = -2.84, R) >consensus ____________GUUGUUU_UGGCCGUUGUUUAUGAAACCGAAUGGUUAAUAAUGGCUAAUUGAAUUCCUGUUGCCGCU_GGCGACAAA_UCAUUUAGCUUG .......................((((((((.....(((......))))))))))).(((.(((.....(((((((....)))))))...))).)))..... (-11.98 = -12.19 + 0.21)

| Location | 8,124,922 – 8,125,021 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Shannon entropy | 0.34383 |
| G+C content | 0.36372 |
| Mean single sequence MFE | -13.16 |
| Consensus MFE | -9.28 |
| Energy contribution | -9.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
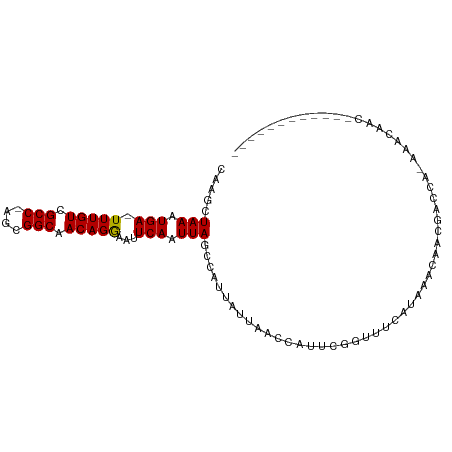
>dm3.chr3R 8124922 99 - 27905053 CAAGCUAAAUGA-UUUGUCGCC-AGCGGCAACAGGAAUUCAAUUAGCCAUUAUUAACUAUUCGGUUUCAUAAACAACGGCCA-AAAAAGAAAAAAAAAAAAC ...(((((.(((-(((((.(((-...))).)))))...))).)))))...............((((...........)))).-................... ( -14.40, z-score = -0.72, R) >droSim1.chr3R 14255332 87 + 27517382 CAAGCUAAAUGA-UUUGUCGCC-AGCGGCAACAGGAAUUCAAUUAGCCAUUAUUAACCAUUCGGUUUCAUAAACAACGGCCA-AAACAAC------------ ...(((((.(((-(((((.(((-...))).)))))...))).)))))..((((.((((....))))..))))..........-.......------------ ( -15.60, z-score = -1.28, R) >droSec1.super_0 7298536 87 + 21120651 CAAGCUAAAUGA-UUUGUCGCC-AGCGGCAACAGGAAUUCAAUUAGACAUUAUUAACCAUUCGGUUUCAUAAACAACGGCCA-AAACAAC------------ ...(((((.(((-(((((.(((-...))).)))))...))).)))).).((((.((((....))))..))))..........-.......------------ ( -12.70, z-score = -0.55, R) >droYak2.chr3R 12442346 87 - 28832112 CAAGCUAAAUGA-UUUGUCGCC-AGCGGCAACAGGAAUUCAAUUAGCCAUUAUUAACCAUUCGGUUUCAUAAACAACGUCCA-AAACAAC------------ ...(((((.(((-(((((.(((-...))).)))))...))).)))))..((((.((((....))))..))))..........-.......------------ ( -15.60, z-score = -1.86, R) >droEre2.scaffold_4770 4255501 87 + 17746568 CAAGCUAAAUGA-UUUGUCGCC-AGCGGCAACAGGAAUUCAAUUAGCCAUUAUUAACCAUUCGGUUUCAUAAACAACGGCCA-AAACAAC------------ ...(((((.(((-(((((.(((-...))).)))))...))).)))))..((((.((((....))))..))))..........-.......------------ ( -15.60, z-score = -1.28, R) >droAna3.scaffold_13340 19416288 88 + 23697760 CAAGUUAACUGA-UUUGUCGCC-GGCGGCAACAGAAAUUCAAUUAGCCAUUAUUAACCAUUCGGCUUCAUAAACAACAACGGCCAACAAC------------ ...(((((.(((-(((((.(((-...))).)))))...))).)))))...............((((..............))))......------------ ( -15.04, z-score = -0.90, R) >droWil1.scaffold_181089 207979 82 - 12369635 CAAGAUAAAUGAUUUUGUCGCCGAAAGGCAACAGAAAUUCAAUUAGCAAUUAUUAACCAUUUCAUUUCAUAGACA--UAACAAG------------------ ...(.(((.(((((((((.(((....))).))))))..))).))).)............................--.......------------------ ( -14.30, z-score = -3.44, R) >droVir3.scaffold_12855 9613482 77 - 10161210 CAAGAUAAAUGA-UUUGUCGCC-AAAGGCAACAGAAAUUCAAUUAGCCAUUAUUAACCAUUUCAUUUCAUAAACAACCA----------------------- .....(((.(((-(((((.(((-...))).)))))...))).)))..................................----------------------- ( -7.50, z-score = -0.91, R) >droGri2.scaffold_14830 2022424 88 - 6267026 CAAGAUAAAUGA-UUUGUCGCC-AAAGGCAACAGAAAUUCAAUUAACCAUUAUUAACCAUUUCAUUUCAUAAGCAACCAAUAACAACAAC------------ ......((((((-(((((.(((-...))).))))).......((((......)))).....)))))).......................------------ ( -7.70, z-score = -1.09, R) >consensus CAAGCUAAAUGA_UUUGUCGCC_AGCGGCAACAGGAAUUCAAUUAGCCAUUAUUAACCAUUCGGUUUCAUAAACAACGACCA_AAACAAC____________ .....(((.(((.(((((.(((....))).)))))...))).)))......................................................... ( -9.28 = -9.03 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:53 2011