
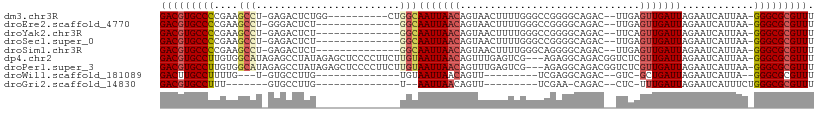
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,123,880 – 8,123,975 |
| Length | 95 |
| Max. P | 0.900107 |

| Location | 8,123,880 – 8,123,975 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.73 |
| Shannon entropy | 0.49855 |
| G+C content | 0.48548 |
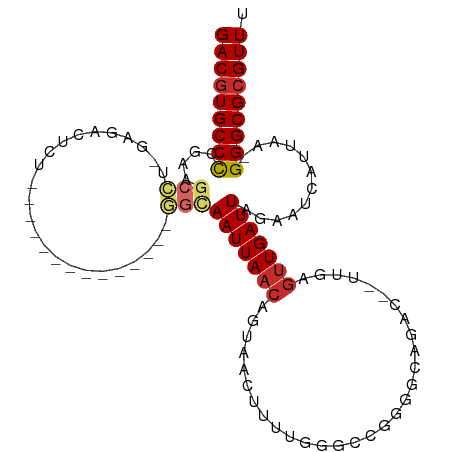
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8123880 95 + 27905053 GACGUGCCCCGAAGCCU-GAGACUCUGG----------CUGGCAAUUAACAGUAACUUUUGGGCCGGGGCAGAC--UUGAGUUGAUUAGAAUCAUUAA-GGGCGCGUUU (((((((((((..(((.-.(((.....(----------(((........))))...)))..)))))))))...(--((((..((((....))))))))-)...))))). ( -34.10, z-score = -2.18, R) >droEre2.scaffold_4770 4254471 91 - 17746568 GACGUGCCCCGAAGCCU-GGGACUCU--------------GGCAAUUAACAGUAACUUUUGGGCCGGGGCAGAC--UUGAGUUGAUUAGAAUCAUUAA-GGGCGCGUUU (((((((((((..(((.-.(((..((--------------(........)))....)))..)))))))))...(--((((..((((....))))))))-)...))))). ( -32.10, z-score = -1.96, R) >droYak2.chr3R 12441305 91 + 28832112 GACGUGCCCCGAAGCCU-GAGACUCU--------------GGCAAUUAACAGUAACUUUUGGGCCGGGGCAGAC--UUCAGUUGAUUAGAAUCAUUAA-GGGCGCGUUU (((((((((.((((.((-(...((((--------------(((......(((......))).)))))))))).)--)))...((((....))))....-))))))))). ( -34.10, z-score = -2.95, R) >droSec1.super_0 7297497 91 - 21120651 GACGUGCCCCGAAGCCU-GAGACUCU--------------GGCAAUUAACAGUAACUUUUGGGCCGGGGCAGAC--UUGAGUUGAUUAGAAUCAUUAA-GGGCGCGUUU (((((((((((..(((.-.(((..((--------------(........)))....)))..)))))))))...(--((((..((((....))))))))-)...))))). ( -32.20, z-score = -2.40, R) >droSim1.chr3R 14254297 91 - 27517382 GACGUGCCCCGAAGCCU-GAGACUCU--------------GGCAAUUAACAGUAACUUUUGGGCAGGGGCAGAC--UUGAGUUGAUUAGAAUCAUUAA-GGGCGCGUUU ((((((((((...(((.-.(((..((--------------(........)))....)))..))).)))))...(--((((..((((....))))))))-)...))))). ( -31.00, z-score = -2.31, R) >dp4.chr2 22911502 105 + 30794189 GACGUGCCUUGUGGCAUAGAGCCUAUAGAGCUCCCCUUCUUGUAAUUAACAGUUUGAGUCG---AGAGGCAGACGGUCUCGUUGAUUAGAAUCAUUAA-GGGCGCGUUU ((((((((((..((....(((((....).)))).))..........(((..((((.(((((---((((((.....))))).)))))).))))..))))-))))))))). ( -31.60, z-score = -1.27, R) >droPer1.super_3 5687166 105 + 7375914 GACGUGCCUUGUGGCAUAGAGCCUAUAGAGCUCCCCUUCUUGUAAUUAACAGUUUGAGUCG---AGAGGCAGACGGUCUCGUUGAUUAGAAUCAUUAA-GGGCGCGUUU ((((((((((..((....(((((....).)))).))..........(((..((((.(((((---((((((.....))))).)))))).))))..))))-))))))))). ( -31.60, z-score = -1.27, R) >droWil1.scaffold_181089 12160975 77 - 12369635 GACUUGCCUUUUG---U-GUGCCUUG--------------UGUAAUUAACAGUU---------UCGAGGCAGAC--GUC-GCUGAUUAGAAUCAUUA--GGGCGCGUUU (((.((((((..(---(-.(((((((--------------...(((.....)))---------.))))))).))--...-..((((....))))..)--))))).))). ( -21.70, z-score = -1.21, R) >droGri2.scaffold_14830 2020795 73 + 6267026 GACGUGCCUUU-------GUGCCUUG--------------U--AAUUAACAGUU---------UCGAA-CAGAC--CUC-UUUGAUUAGAAUCAUUUCUGGGCGCGUUU (((((((((..-------(((..((.--------------(--((((((.((..---------((...-..)).--..)-)))))))).)).)))....))))))))). ( -19.40, z-score = -2.18, R) >consensus GACGUGCCCCGAAGCCU_GAGACUCU______________GGCAAUUAACAGUAACUUUUGGGCCGGGGCAGAC__UUGAGUUGAUUAGAAUCAUUAA_GGGCGCGUUU (((((((((....(((........................)))(((((((..............................)))))))............))))))))). (-13.86 = -14.08 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:51 2011