
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,114,360 – 8,114,483 |
| Length | 123 |
| Max. P | 0.771870 |

| Location | 8,114,360 – 8,114,458 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.60 |
| Shannon entropy | 0.17020 |
| G+C content | 0.49834 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8114360 98 + 27905053 UGCUCAUCCCGAAAAGUUAAGAUACCCAGAGAUACUCGGGAUACUGGGAUGGGAUGGGUUGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCA .((((((((((...(((....((.(((.(((...)))))))))))....))))))))))(((((((((....)))))))))....((.(....).)). ( -34.90, z-score = -2.50, R) >droSim1.chr3R 14243629 93 - 27517382 UGUUCAUCCCGAAAAGUUAAGAUACCCAGAGAUACUCGGGAUACUGGGAUGGGAUG-----CAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCA ..(((((((((....((....)).(((.(((...))))))....))))))))).((-----(((((((....)))))))))....((.(....).)). ( -29.40, z-score = -1.86, R) >droSec1.super_0 7288099 93 - 21120651 UGUUCAUCCCGAAAAGUUAAGAUACCCAGAGAUACUCGGGAUACUGGGAUGGGAUG-----CAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCA ..(((((((((....((....)).(((.(((...))))))....))))))))).((-----(((((((....)))))))))....((.(....).)). ( -29.40, z-score = -1.86, R) >droYak2.chr3R 12431765 83 + 28832112 CGCUCAUCCCGAAAAGUUAAGAUACCCAGA--UACUCGGGAU--------GGGAUG-----CAGCUCUAGAUAGAGCUGCGAUAGGCGCAUAUGGGCA ..((((((((((..................--...)))))))--------))).((-----(((((((....)))))))))....((.(....).)). ( -32.60, z-score = -3.11, R) >droEre2.scaffold_4770 4244962 85 - 17746568 UGUUCAUCCCGAGAAGUUAAGAUACCCAGAGAUACUCGGGAU--------GGGAUG-----CAGCUCCAGAUAGAGUUGCGAUAGGCGCAUAUGGGCA ..(((((((((((...((..(.....)..))...))))))))--------))).((-----((((((......))))))))....((.(....).)). ( -29.20, z-score = -2.45, R) >consensus UGUUCAUCCCGAAAAGUUAAGAUACCCAGAGAUACUCGGGAUACUGGGAUGGGAUG_____CAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCA .(((((((((.....((....)).(((.(((...))))))..........))))).......((((((....))))))(((.....)))....)))). (-21.80 = -21.80 + 0.00)



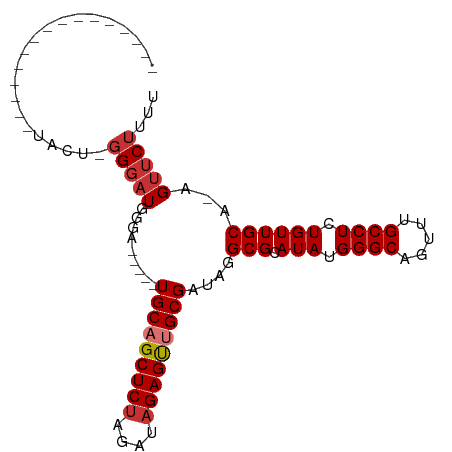
| Location | 8,114,392 – 8,114,483 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.35818 |
| G+C content | 0.51740 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8114392 91 + 27905053 -------UACUCGGGAUACU-GGGAUGGGAUGGGUUGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCA-AGUUCUUUU -------..(((((....))-)))..(((((..(((((((((((....)))))))))))..(((.(((.((((.....)))).)))))).-.)))))... ( -32.00, z-score = -2.17, R) >droSim1.chr3R 14243661 86 - 27517382 -------UACUCGGGAUACU-GGGAUGGGA-----UGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCA-AGUUCUUUU -------..(((((....))-)))..((((-----(((((((((....)))))))).....(((.(((.((((.....)))).)))))).-.)))))... ( -29.00, z-score = -1.69, R) >droSec1.super_0 7288131 86 - 21120651 -------UACUCGGGAUACU-GGGAUGGGA-----UGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCA-AGUUCUUUU -------..(((((....))-)))..((((-----(((((((((....)))))))).....(((.(((.((((.....)))).)))))).-.)))))... ( -29.00, z-score = -1.69, R) >droYak2.chr3R 12431795 78 + 28832112 ----------------UACUCGGGAUGGGA-----UGCAGCUCUAGAUAGAGCUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCA-AGUUCUUUU ----------------.....(((((....-----(((((((((....)))))))))....(((.(((.((((.....)))).)))))).-.)))))... ( -26.30, z-score = -1.07, R) >droEre2.scaffold_4770 4244994 78 - 17746568 ----------------UACUCGGGAUGGGA-----UGCAGCUCCAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCG-AGUUCUUUU ----------------.(((((.(((((..-----((((((((......))))))))............((((.....))))))))).))-)))...... ( -26.70, z-score = -1.29, R) >droAna3.scaffold_13340 19406484 97 - 23697760 UACUCGGAUACUUGGAUGCU-CGCCUGCUCGAAGCUGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGU-UGCCUCUGUUGCA-AGUUCUUUU .....(((.(((((.((((.-.((((((.....))(((((((((....)))))))))..))))))))..((((...-.))))......))-))))))... ( -32.10, z-score = -0.38, R) >dp4.chr2 22900791 80 + 30794189 -------------------UGGGGAUGGGGCGG-CUGCAGCUCUAGAUAGAGUCGCGAUAGGCGCAUAUGGGCAGUGUGCCUCUGUUGCACAGUUCUUUC -------------------.((((((...((((-((((.(((((....))))).)))..((((((((.......))))))))..)))))...)))))).. ( -29.20, z-score = -0.79, R) >droPer1.super_3 5676417 80 + 7375914 -------------------UGGGGAUGGGGCGG-CUGCAGCUCUAGAUAGAGUCGCGAUAGGCGCAUAUGGGCAGUGUGCCUCUGUUGCACAGUUCUUUC -------------------.((((((...((((-((((.(((((....))))).)))..((((((((.......))))))))..)))))...)))))).. ( -29.20, z-score = -0.79, R) >consensus ________________UACU_GGGAUGGGA_____UGCAGCUCUAGAUAGAGUUGCGAUAGGCGCAUAUGGGCAGUUUGCCUCUGUUGCA_AGUUCUUUU .....................(((((.........(((((((((....)))))))))....(((.(((.((((.....)))).))))))...)))))... (-21.40 = -22.04 + 0.64)

| Location | 8,114,392 – 8,114,483 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Shannon entropy | 0.35818 |
| G+C content | 0.51740 |
| Mean single sequence MFE | -19.41 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.58 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
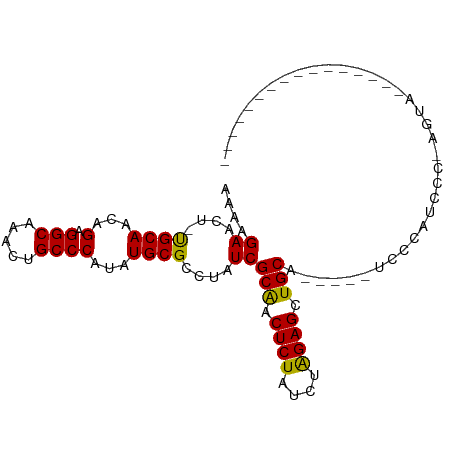
>dm3.chr3R 8114392 91 - 27905053 AAAAGAACU-UGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUAGAGCUGCAACCCAUCCCAUCCC-AGUAUCCCGAGUA------- ......(((-((((...(.(((.....))))...))).......(((.((((....)))).)))...............-........)))).------- ( -16.80, z-score = -1.28, R) >droSim1.chr3R 14243661 86 + 27517382 AAAAGAACU-UGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUAGAGCUGCA-----UCCCAUCCC-AGUAUCCCGAGUA------- ......(((-((((...(.(((.....))))...))).......(((.((((....)))).))).-----.........-........)))).------- ( -16.80, z-score = -0.99, R) >droSec1.super_0 7288131 86 + 21120651 AAAAGAACU-UGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUAGAGCUGCA-----UCCCAUCCC-AGUAUCCCGAGUA------- ......(((-((((...(.(((.....))))...))).......(((.((((....)))).))).-----.........-........)))).------- ( -16.80, z-score = -0.99, R) >droYak2.chr3R 12431795 78 - 28832112 AAAAGAACU-UGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAGCUCUAUCUAGAGCUGCA-----UCCCAUCCCGAGUA---------------- ......(((-((((...(.(((.....))))...))).......((((((((....)))))))).-----.........)))).---------------- ( -23.40, z-score = -2.99, R) >droEre2.scaffold_4770 4244994 78 + 17746568 AAAAGAACU-CGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUGGAGCUGCA-----UCCCAUCCCGAGUA---------------- ......(((-((((...(.(((.....))))...))).......(((.((((....)))).))).-----.........)))).---------------- ( -19.70, z-score = -1.75, R) >droAna3.scaffold_13340 19406484 97 + 23697760 AAAAGAACU-UGCAACAGAGGCA-ACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUAGAGCUGCAGCUUCGAGCAGGCG-AGCAUCCAAGUAUCCGAGUA ......(((-((..((.(.(((.-...)))).....(((((...(((.((((....)))).))).((.....)))))))-.........))...))))). ( -26.00, z-score = -0.38, R) >dp4.chr2 22900791 80 - 30794189 GAAAGAACUGUGCAACAGAGGCACACUGCCCAUAUGCGCCUAUCGCGACUCUAUCUAGAGCUGCAG-CCGCCCCAUCCCCA------------------- ....((...(((((...(.(((.....))))...)))))...))(((.((((....)))).)))..-..............------------------- ( -17.90, z-score = -0.45, R) >droPer1.super_3 5676417 80 - 7375914 GAAAGAACUGUGCAACAGAGGCACACUGCCCAUAUGCGCCUAUCGCGACUCUAUCUAGAGCUGCAG-CCGCCCCAUCCCCA------------------- ....((...(((((...(.(((.....))))...)))))...))(((.((((....)))).)))..-..............------------------- ( -17.90, z-score = -0.45, R) >consensus AAAAGAACU_UGCAACAGAGGCAAACUGCCCAUAUGCGCCUAUCGCAACUCUAUCUAGAGCUGCA_____UCCCAUCCC_AGUA________________ ....((....((((...(.(((.....))))...))))....))(((.((((....)))).))).................................... (-15.98 = -15.58 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:51 2011