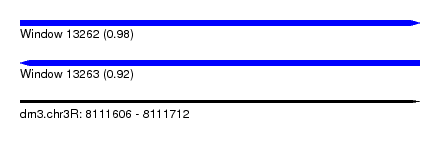
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,111,606 – 8,111,712 |
| Length | 106 |
| Max. P | 0.976370 |

| Location | 8,111,606 – 8,111,712 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 61.81 |
| Shannon entropy | 0.64343 |
| G+C content | 0.40159 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -10.67 |
| Energy contribution | -10.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
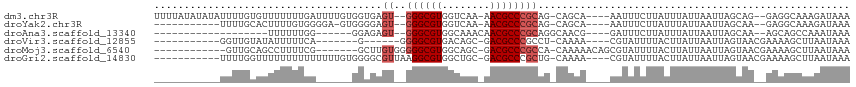
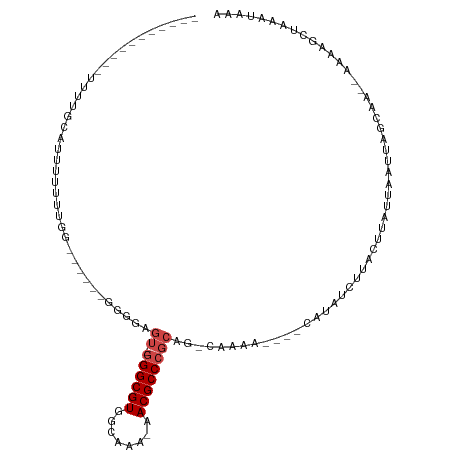
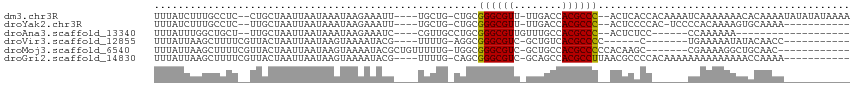
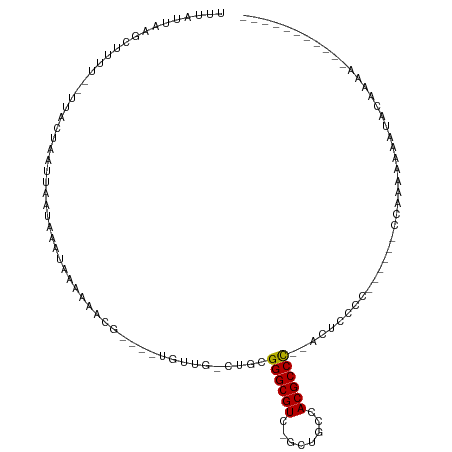
>dm3.chr3R 8111606 106 + 27905053 UUUUAUAUAUAUUUUGUGUUUUUUUGAUUUUGUGGUGAGU--GGGCGUGGUCAA-AACGCCCGCAG-CAGCA----AAUUUCUUAUUUAUUAAUUAGCAG--GAGGCAAAGAUAAA ...........((((((.((((.((((((....((((((.--((((((......-.))))))((..-..)).----.....))))))....)))))).))--)).))))))..... ( -26.80, z-score = -1.99, R) >droYak2.chr3R 12429003 94 + 28832112 -----------UUUUGCACUUUUGUGGGGA-GUGGGGAGU--GGGCGUGGUCAA-AACGCCCGCAG-CAGCA----AAUUUCUUAUUUAUUAAUUAGCAA--GAGGCAAAGAUAAA -----------((((((.(((((((...((-(((((((((--((((((......-.))))))((..-..)).----.)))))))))))........))))--)))))))))..... ( -33.60, z-score = -3.98, R) >droAna3.scaffold_13340 19403957 83 - 23697760 -------------------UUUUUUGG------GGAGAGU--GGGCGUGGCAAACAACGCCCGCAGGCAACG----GAUUUCUUAUUUAUUAAUUAGCAA--AGCAGCCAAAUAAA -------------------...(((((------(((((((--((((((........)))))))).(....).----..))))).............((..--.))..))))).... ( -23.80, z-score = -1.89, R) >droVir3.scaffold_12855 9590301 86 + 10161210 -----------GGUUGUAUAUUUUUCA-------G------GGGGCGUGACAGC-GACGCCCGCCU-CAAAA----CGUAUUUUACUUAUUAAUUAGUAACGAAAAGCUUAAUAAA -----------.....((..(((((((-------(------(((((((......-.)))))).)))-.....----......(((((........))))).))))))..))..... ( -19.60, z-score = -1.04, R) >droMoj3.scaffold_6540 24653558 95 + 34148556 ------------GUUGCAGCCUUUUCG-------GCUUGUGGGGGCGUGGCAGC-GACGCCCGCCA-CAAAAACAGCGUAUUUUACUUAUUAAUUAGUAACGAAAAGCUUAAUAAA ------------((((.(((.((((((-------((((((((((((((.(...)-.)))))).)))-)).....))).....(((((........))))))))))))))))))... ( -32.70, z-score = -2.55, R) >droGri2.scaffold_14830 2003983 99 + 6267026 -----------UUUUGGUUUUUUUUUUUUUUGUGGGGCGUUAAGGCGUGGCUGC-GACGCCCGCUG-CAAAA----CGUAUUUUACUUAUUAAUUAGUAACGAAAAGCUUAAUAAA -----------....((((((((....((((((((((((((..(((...)))..-))))))).).)-)))))----......(((((........))))).))))))))....... ( -24.80, z-score = -0.90, R) >consensus ___________UUUUGCAUUUUUUUGG______GGGGAGU__GGGCGUGGCAAA_AACGCCCGCAG_CAAAA____CAUAUCUUACUUAUUAAUUAGCAA__AAAAGCUAAAUAAA ..........................................((((((........))))))...................................................... (-10.67 = -10.83 + 0.17)
| Location | 8,111,606 – 8,111,712 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 61.81 |
| Shannon entropy | 0.64343 |
| G+C content | 0.40159 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8111606 106 - 27905053 UUUAUCUUUGCCUC--CUGCUAAUUAAUAAAUAAGAAAUU----UGCUG-CUGCGGGCGUU-UUGACCACGCCC--ACUCACCACAAAAUCAAAAAAACACAAAAUAUAUAUAAAA ......((((....--........................----.((..-..))((((((.-......))))))--........))))............................ ( -12.90, z-score = -0.34, R) >droYak2.chr3R 12429003 94 - 28832112 UUUAUCUUUGCCUC--UUGCUAAUUAAUAAAUAAGAAAUU----UGCUG-CUGCGGGCGUU-UUGACCACGCCC--ACUCCCCAC-UCCCCACAAAAGUGCAAAA----------- ......(((((((.--(((.....................----.((..-..))((((((.-......))))))--.........-......))).)).))))).----------- ( -19.20, z-score = -1.98, R) >droAna3.scaffold_13340 19403957 83 + 23697760 UUUAUUUGGCUGCU--UUGCUAAUUAAUAAAUAAGAAAUC----CGUUGCCUGCGGGCGUUGUUUGCCACGCCC--ACUCUCC------CCAAAAAA------------------- ....(((((..((.--..((....................----....))..))((((((.(....).))))))--.......------)))))...------------------- ( -17.46, z-score = -0.69, R) >droVir3.scaffold_12855 9590301 86 - 10161210 UUUAUUAAGCUUUUCGUUACUAAUUAAUAAGUAAAAUACG----UUUUG-AGGCGGGCGUC-GCUGUCACGCCCC------C-------UGAAAAAUAUACAACC----------- ................(((((........)))))......----((((.-(((.((((((.-......)))))))------)-------).))))..........----------- ( -19.20, z-score = -1.60, R) >droMoj3.scaffold_6540 24653558 95 - 34148556 UUUAUUAAGCUUUUCGUUACUAAUUAAUAAGUAAAAUACGCUGUUUUUG-UGGCGGGCGUC-GCUGCCACGCCCCCACAAGC-------CGAAAAGGCUGCAAC------------ .......((((((((((((((........)))))...........((((-(((.((((((.-......))))))))))))).-------)))))).))).....------------ ( -29.70, z-score = -2.27, R) >droGri2.scaffold_14830 2003983 99 - 6267026 UUUAUUAAGCUUUUCGUUACUAAUUAAUAAGUAAAAUACG----UUUUG-CAGCGGGCGUC-GCAGCCACGCCUUAACGCCCCACAAAAAAAAAAAAAACCAAAA----------- ........((....(((((((........))))....)))----....)-).(.((((((.-((......))....)))))))......................----------- ( -16.40, z-score = -0.41, R) >consensus UUUAUUAAGCUUUU__UUACUAAUUAAUAAAUAAAAAACG____UGUUG_CUGCGGGCGUC_GCUGCCACGCCC__ACUCCCC______CCAAAAAAAUACAAAA___________ ......................................................((((((........)))))).......................................... ( -9.54 = -9.40 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:49 2011