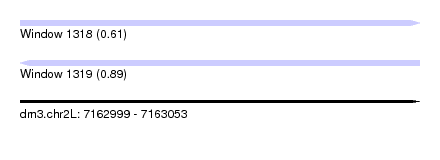
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,162,999 – 7,163,053 |
| Length | 54 |
| Max. P | 0.887550 |

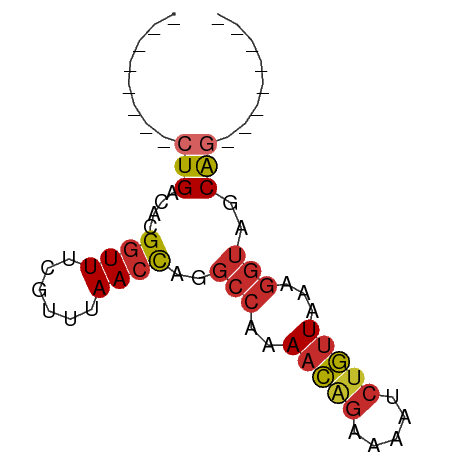
| Location | 7,162,999 – 7,163,053 |
|---|---|
| Length | 54 |
| Sequences | 9 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Shannon entropy | 0.35868 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -12.39 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7162999 54 + 23011544 --------CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG-------- --------(((.(..((((......))))..(((..(((((.....)))))...))).))))-------- ( -12.10, z-score = -1.75, R) >droPer1.super_8 866754 62 - 3966273 --------CUGACAUGGUUUCGCUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCGCACAGACAG --------(((...(((((......))))).(((..(((((.....)))))...)))......))).... ( -14.00, z-score = -1.43, R) >dp4.chr4_group2 802102 62 + 1235136 --------CUGACAUGGUUUCGCUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCGCACAGACAG --------(((...(((((......))))).(((..(((((.....)))))...)))......))).... ( -14.00, z-score = -1.43, R) >droAna3.scaffold_12943 1012005 61 - 5039921 CUGAAAACCUGGCAUGGUUUCG-UUAACCAGGCCAGAACGGAAAAUCUGUUAAAGGUAACGG-------- (((.....(((((.(((((...-..))))).)))))(((((.....)))))........)))-------- ( -19.10, z-score = -2.76, R) >droEre2.scaffold_4929 16078132 54 + 26641161 --------CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG-------- --------(((.(..((((......))))..(((..(((((.....)))))...))).))))-------- ( -12.10, z-score = -1.75, R) >droYak2.chr2L 16581811 54 - 22324452 --------CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG-------- --------(((.(..((((......))))..(((..(((((.....)))))...))).))))-------- ( -12.10, z-score = -1.75, R) >droSec1.super_3 2686624 54 + 7220098 --------CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG-------- --------(((.(..((((......))))..(((..(((((.....)))))...))).))))-------- ( -12.10, z-score = -1.75, R) >droSim1.chr2L 6952862 54 + 22036055 --------CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG-------- --------(((.(..((((......))))..(((..(((((.....)))))...))).))))-------- ( -12.10, z-score = -1.75, R) >droMoj3.scaffold_6500 27979399 54 - 32352404 --------CUGACAUGGUUUCGUUUAAGUGGCGCAGAGUGAAAAAAAAAAUACAAAUAUCAG-------- --------((((.((.((((..(((...(.((.....)).).)))..))))....)).))))-------- ( -3.90, z-score = 1.05, R) >consensus ________CUGACACGGUUUCGUUUAACCAGGCCAAAACAGAAAAUCUGUUAAAGGUAGCAG________ ........(((....((((......))))..(((..(((((.....)))))...)))..)))........ ( -8.14 = -8.53 + 0.40)

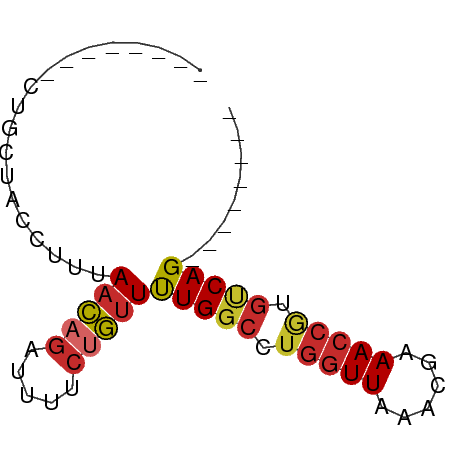
| Location | 7,162,999 – 7,163,053 |
|---|---|
| Length | 54 |
| Sequences | 9 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Shannon entropy | 0.35868 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -11.88 |
| Consensus MFE | -9.75 |
| Energy contribution | -10.11 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7162999 54 - 23011544 --------CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG-------- --------...........(((((.....))))).((((.(((((......))))).)))).-------- ( -12.00, z-score = -1.68, R) >droPer1.super_8 866754 62 + 3966273 CUGUCUGUGCGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAGCGAAACCAUGUCAG-------- ..((((((............)))))).........((((.(((((......))))).)))).-------- ( -13.80, z-score = -0.97, R) >dp4.chr4_group2 802102 62 - 1235136 CUGUCUGUGCGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAGCGAAACCAUGUCAG-------- ..((((((............)))))).........((((.(((((......))))).)))).-------- ( -13.80, z-score = -0.97, R) >droAna3.scaffold_12943 1012005 61 + 5039921 --------CCGUUACCUUUAACAGAUUUUCCGUUCUGGCCUGGUUAA-CGAAACCAUGCCAGGUUUUCAG --------..((((....))))...........((((((.(((((..-...))))).))))))....... ( -16.00, z-score = -2.55, R) >droEre2.scaffold_4929 16078132 54 - 26641161 --------CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG-------- --------...........(((((.....))))).((((.(((((......))))).)))).-------- ( -12.00, z-score = -1.68, R) >droYak2.chr2L 16581811 54 + 22324452 --------CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG-------- --------...........(((((.....))))).((((.(((((......))))).)))).-------- ( -12.00, z-score = -1.68, R) >droSec1.super_3 2686624 54 - 7220098 --------CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG-------- --------...........(((((.....))))).((((.(((((......))))).)))).-------- ( -12.00, z-score = -1.68, R) >droSim1.chr2L 6952862 54 - 22036055 --------CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG-------- --------...........(((((.....))))).((((.(((((......))))).)))).-------- ( -12.00, z-score = -1.68, R) >droMoj3.scaffold_6500 27979399 54 + 32352404 --------CUGAUAUUUGUAUUUUUUUUUUCACUCUGCGCCACUUAAACGAAACCAUGUCAG-------- --------(((((((((((............................))))....)))))))-------- ( -3.29, z-score = 0.21, R) >consensus ________CUGCUACCUUUAACAGAUUUUCUGUUUUGGCCUGGUUAAACGAAACCGUGUCAG________ ...................(((((.....)))))(((((.(((((......))))).)))))........ ( -9.75 = -10.11 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:03 2011