
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,095,425 – 8,095,529 |
| Length | 104 |
| Max. P | 0.749063 |

| Location | 8,095,425 – 8,095,529 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Shannon entropy | 0.31692 |
| G+C content | 0.45175 |
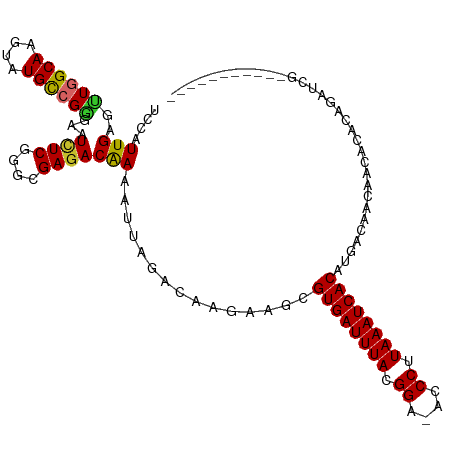
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.50 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
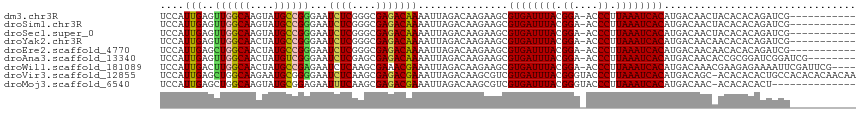
>dm3.chr3R 8095425 104 + 27905053 UCCAUUGAGUUGGCAAGUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACUACACACAGAUCG----------- .....(((((((.((.((.((((.((....)).))))..)).................((((((((.((.-..)).)))))))).)).))))).)).........----------- ( -22.20, z-score = -0.83, R) >droSim1.chr3R 14224600 104 - 27517382 UCCAUUGAGUUGGCAAGUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACUACACACAGAUCG----------- .....(((((((.((.((.((((.((....)).))))..)).................((((((((.((.-..)).)))))))).)).))))).)).........----------- ( -22.20, z-score = -0.83, R) >droSec1.super_0 7269108 104 - 21120651 UCCAUUGAGUUGGCAAGUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACUACACACAGAUCG----------- .....(((((((.((.((.((((.((....)).))))..)).................((((((((.((.-..)).)))))))).)).))))).)).........----------- ( -22.20, z-score = -0.83, R) >droYak2.chr3R 12412794 104 + 28832112 UCCAUUGAGUUGGCAACUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACAACACACAGAUCG----------- ........((((.((.((.((((.((....)).))))))...................((((((((.((.-..)).)))))))).)).)))).............----------- ( -21.30, z-score = -0.93, R) >droEre2.scaffold_4770 4226057 104 - 17746568 UCCAUUGAGCUGGCAACUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACAACACACAGAUCG----------- ....(((..((((((....))))))...((((....)))))))...............((((((((.((.-..)).)))))))).....................----------- ( -22.10, z-score = -1.35, R) >droAna3.scaffold_13340 19387876 107 - 23697760 UCCAUUGAGUUGGCAACUAUGUCGGGAAUCUCGAGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAACACCGCGGAUCGGAUCG-------- ......((((((((..((.((((.....((((....))))........))))))..))((((((((.((.-..)).))))))))....)))).(((.....))).)).-------- ( -25.92, z-score = -0.90, R) >droWil1.scaffold_181089 240895 111 + 12369635 UCCAUUGACUUGGCAACUAUGCCGAGAAUCUCAAGCGAAACGAAAUUAGACAAGAAGCGUGAUUUACGGA-ACCCUUAAAUCACAUGACAAACGAAGAGAAAAUUCGAUUCG---- ...((((((((((((....)))))))..((((..((....................))((((((((.((.-..)).))))))))............))))....)))))...---- ( -26.05, z-score = -3.03, R) >droVir3.scaffold_12855 9565265 115 + 10161210 UCCAUUGAGCUGGCAAGAAUGCGGGGAAUCUCAAGCGAGACGAAAUUAGACAAGCGUCGUGAUUUACGGGUACCCUUAAAUCACAUGACAGC-ACACACACUGCCACACACAACAA ........(.(((((.......(((....)))..((..((((............))))((((((((.((....)).))))))))......))-........))))))......... ( -26.70, z-score = -1.11, R) >droMoj3.scaffold_6540 24631694 101 + 34148556 UCCAUUGAGCUGGCAAGUAUGCGGAGAAUUUCAAGCGAGACGAAAUUAGACAAGCGUCGUGAUUUACGGGUACCCUUAAAUCACAUGACAAC-ACACACACU-------------- ........(((.(((....)))(...((((((.........))))))...).)))(((((((((((.((....)).))))))))..)))...-.........-------------- ( -22.20, z-score = -1.13, R) >consensus UCCAUUGAGUUGGCAAGUAUGCCGGGAAUCUCGGGCGAGACAAAAUUAGACAAGAAGCGUGAUUUACGGA_ACCCUUAAAUCACAUGACAACAACACACAGAUCG___________ ....(((..((((((....))))))...((((....)))))))...............((((((((.((....)).))))))))................................ (-19.70 = -19.50 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:43 2011