| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,091,360 – 8,091,451 |
| Length | 91 |
| Max. P | 0.969023 |

| Location | 8,091,360 – 8,091,451 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 67.34 |
| Shannon entropy | 0.68474 |
| G+C content | 0.52181 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -9.63 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
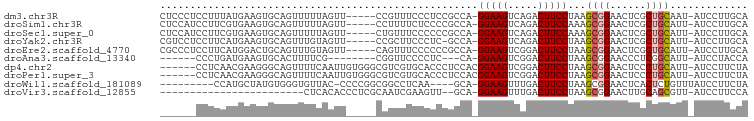
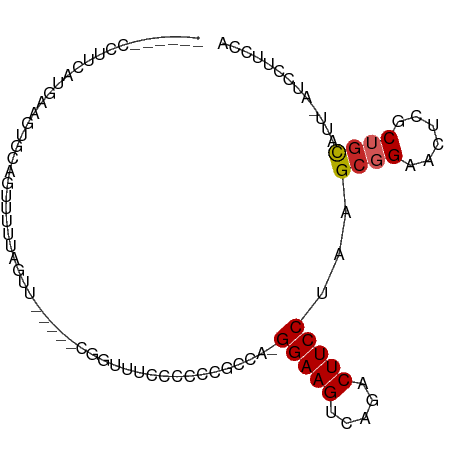
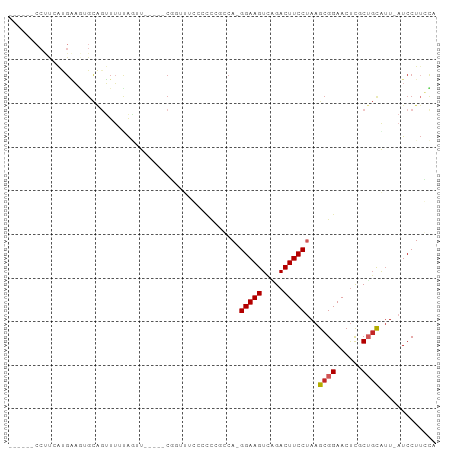
>dm3.chr3R 8091360 91 + 27905053 CUCCCUCCUUUAUGAAGUGCAGUUUUUAGUU-----CCGUUUCCCUCCGCCA-GGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAAU-AUCCUUGCA .............((..((((((....((((-----((((((.........(-(((((.....)))))))))))))))).))))))..-.))...... ( -26.70, z-score = -3.61, R) >droSim1.chr3R 14220592 91 - 27517382 CUCCAUCCUUCGUGAAGUGCAGUUUUUAGUU-----CCUUUUCUCCCCGCCA-GGAAGUCAGACUUCCAAAGCGGAACUCGCUGCAUU-AUCCUUGCA ...........(..(((((((((....((((-----((..........((..-(((((.....)))))...)))))))).))))))))-....)..). ( -22.90, z-score = -2.15, R) >droSec1.super_0 7265132 91 - 21120651 CUCCAUCCUUCGUGAAGUGCAGUUUUUAGUU-----CUGUUUCCCCCCGCCA-GGAAGUCAGACUUCCAAAGCGGAACUCGCUGCAUU-AUCCUUGCA ...........(..(((((((((....((((-----((((((..........-(((((.....))))).)))))))))).))))))))-....)..). ( -24.60, z-score = -2.54, R) >droYak2.chr3R 12408672 90 + 28832112 CGUCCUCCUUCAUGAAGUGCAGUUUGUAGUU-----CCGCUUCCCUC-GCCA-GGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAUU-AUCCUUGCA ...............((((((((..(.((((-----((((((.....-...(-(((((.....)))))))))))))))))))))))))-......... ( -31.10, z-score = -4.07, R) >droEre2.scaffold_4770 4222107 91 - 17746568 CGCCCUCCUUCAUGGACUGCAGUUUGUAGUU-----CAGUUUCCCCCCGCCA-GGAAGUCGGACUUCCUAAGCGGAACUCGCUGCAUU-AUCCUUGCA .((.........((((((((.....))))))-----))........((((.(-(((((.....))))))..)))).....)).(((..-.....))). ( -27.80, z-score = -2.06, R) >droAna3.scaffold_13340 19384109 79 - 23697760 ------CCCUGAUGAAGUGCACUUUUCG--------CGGUUCCCCUC---CA-GGAAGUCGGACUUCCUAAGCGGAACCCUCGGCAUU-AUCCUACCA ------....(((((.((((.......)--------)((((((.((.---.(-(((((.....)))))).)).))))))....)).))-)))...... ( -23.40, z-score = -1.52, R) >dp4.chr2 22875533 91 + 30794189 ------CCUCAACGAAGGGCAGUUUUCAAUUGUGGGCGUCGUGCACCCUCCACGGAAGUCGGACUUCCUAAGCGGAACUCCCUGCAUU-AUCCUUCUA ------.......(((((((((.((((....(.(((.(.....).))).)....))))..(((.((((.....)))).)))))))...-..))))).. ( -24.50, z-score = -0.24, R) >droPer1.super_3 5651023 91 + 7375914 ------CCUCAACGAAGGGCAGUUUUCAAUUGUGGGCGUCGUGCACCCUCCACGGAAGUCGGACUUCCUAAGCGGAACUCCCUGCAUU-AUCCUUCUA ------.......(((((((((.((((....(.(((.(.....).))).)....))))..(((.((((.....)))).)))))))...-..))))).. ( -24.50, z-score = -0.24, R) >droWil1.scaffold_181089 12124152 83 - 12369635 ---------CCAUGCUAUGUGGGUGUUAC-CCCCGGCGGCCUCAA----GCA-GGAAGUUUGACUUCCUAAGCGGAACUCACUCUGUUUAUCCUUCUA ---------...((((.((.((.((((..-....)))).)).)))----)))-(((((.....)))))((((((((......))))))))........ ( -24.20, z-score = -1.07, R) >droVir3.scaffold_12855 9559967 70 + 10161210 ------------------------CUCACACCCUCGCAAUCGAAGUU--GCA-GGAAGUUUGACUUCCUAAGCGGAACUUGCAGCGUU-AUCCUUCCA ------------------------...........(((((....)))--)).-(((((..((((((((.....))))........)))-)..))))). ( -16.90, z-score = -1.16, R) >consensus ______CCUUCAUGAAGUGCAGUUUUUAGUU_____CGGUUUCCCCCCGCCA_GGAAGUCAGACUUCCUAAGCGGAACUCGCUGCAUU_AUCCUUCCA .....................................................(((((.....)))))...((((......))))............. ( -9.63 = -9.84 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:40 2011