| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,079,568 – 8,079,658 |
| Length | 90 |
| Max. P | 0.951088 |

| Location | 8,079,568 – 8,079,658 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.24 |
| Shannon entropy | 0.53992 |
| G+C content | 0.41655 |
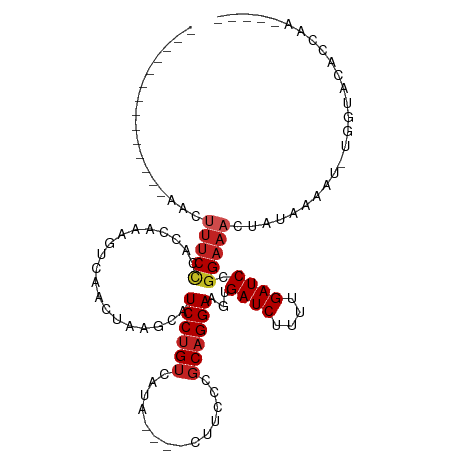
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -11.00 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8079568 90 - 27905053 --------------AACUUUCCCACCAAAGUCAACUAAGCAUCCUGUCAUA----CUUCCCGCAGGAUGUGAUCUUUUGAUCCGGAAACUAUAAAAU-UGGUACACCAA----- --------------...(((((................(((((((((....----......)))))))))((((....)))).)))))........(-(((....))))----- ( -21.80, z-score = -3.13, R) >droPer1.super_3 5638136 98 - 7375914 --------------AACUUUCCCACCAAAUC--GUUGAGCAUCCUGUCAUACUAUUUUCCUGCAGGAAGUGAUCUUUUGAUCCGGAAACUAUAAACUGUGGUACGGCACACACA --------------...(((((.........--.....((.((((((..............)))))).))((((....)))).)))))........((((........)))).. ( -17.94, z-score = 0.19, R) >dp4.chr2 22862624 98 - 30794189 --------------AACUUUCCCACCAAAUC--GUUGAGCAUCCUGUCAUACUAUUUUCCUGCAGGAAGUGAUCUUUUGAUCCGGAAACUAUAAACUGUGGUACGGCACACACA --------------...(((((.........--.....((.((((((..............)))))).))((((....)))).)))))........((((........)))).. ( -17.94, z-score = 0.19, R) >droAna3.scaffold_13340 19372126 89 + 23697760 ---------------ACUUUCCCACCAAAGCCAACCGAGCAUCCAGUCAUA----UUCCCGGCAGGAAAUGAUCUUUUGAUCCGGAAACUAUAAAAU-UGGUACACCGA----- ---------------........(((((.(((....).)).(((.......----((((.....))))..((((....)))).)))..........)-)))).......----- ( -15.10, z-score = -0.02, R) >droEre2.scaffold_4770 4206904 89 + 17746568 ---------------ACUUUCCCACCAAAGUCAAAUAAGCAUCCUGUCAUA----UUUCCCGCAGGAUGUGAUCUUUUGAUCCGGAAACUAUAAAAC-UGGUACACCAA----- ---------------........((((..((((((...(((((((((....----......))))))))).....))))))..(....)........-)))).......----- ( -21.60, z-score = -3.12, R) >droYak2.chr3R 12396667 90 - 28832112 --------------AACUUUCCCACCAAAGUCAACUAAGCAUCCUGUCAUA----UUUCCCGCAGGAUGUGAUCUUUUGAUCCGGAAACUAUAAAAU-UGGUACACCAA----- --------------...(((((................(((((((((....----......)))))))))((((....)))).)))))........(-(((....))))----- ( -21.80, z-score = -3.09, R) >droSec1.super_0 7226462 90 + 21120651 --------------AACUUUCCCACCAAAGUCAACUAAGCAUCCUGUCAUA----CUUCCCGCAGGAUGUGAUCUUUUGAUCCGGAAACUAUAAAAU-UUUUACCCCAA----- --------------...(((((................(((((((((....----......)))))))))((((....)))).))))).........-...........----- ( -19.40, z-score = -3.64, R) >droSim1.chr3R 14208365 90 + 27517382 --------------AACUUUCCCACCAAAGUCAACUAAGCAUCCUGUCAUA----CUUCCCGCAGGAUGUGAUCUUUUGAUCCGGAAACUAUAAAAU-UGGUACACCAA----- --------------...(((((................(((((((((....----......)))))))))((((....)))).)))))........(-(((....))))----- ( -21.80, z-score = -3.13, R) >droGri2.scaffold_14830 1963513 103 - 6267026 AAUGUUCCCACAAGCCCACUCUCAGGCACAGUCUGCAAGCAUCCUGUCAUAC--UCCUUGAGCAGGAAAUGAUCUUUUGAUCCGGAAACAAUAAAAU-U-GUAUAUG------- ..((((.((....(((........)))..............((((((((...--....))).)))))...((((....)))).)).)))).......-.-.......------- ( -20.70, z-score = -0.61, R) >droVir3.scaffold_12855 616229 103 + 10161210 AAUUUUCCCACAAGCCCAUGCAUAUGCACAGCCUGCAAGCAUCCUGUCA-AU--GCCUGGCGCAGGAAAUGAUCUUUUGAUCCGGAAACUAUAAAAU-UGGUUUGUG------- ...(((((.....(((((.((((.((.((((..((....))..))))))-))--)).))).)).))))).((((....))))((.((((((......-)))))).))------- ( -27.10, z-score = -1.10, R) >droWil1.scaffold_181089 269034 80 - 12369635 --------------AAUUUUCCCACCAGUU---AGUAAGCAUCCUGU---------------CAGGAAAUGAUCUUUUGAUCCGGAAACUAUAAAAAUUGGUACACCAACAC-- --------------.........(((((((---..((((..(((.((---------------(((((.......)))))))..)))..)).))..)))))))..........-- ( -13.70, z-score = -0.61, R) >consensus ______________AACUUUCCCACCAAAGUCAACUAAGCAUCCUGUCAUA____CUUCCCGCAGGAAGUGAUCUUUUGAUCCGGAAACUAUAAAAU_UGGUACACCAA_____ .................(((((...................((((((..............))))))...((((....)))).))))).......................... (-11.00 = -11.56 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:39 2011