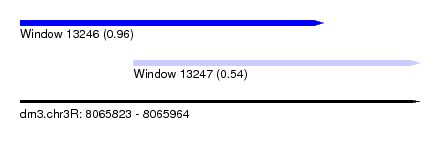
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,065,823 – 8,065,964 |
| Length | 141 |
| Max. P | 0.957593 |

| Location | 8,065,823 – 8,065,930 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.32971 |
| G+C content | 0.53664 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -25.32 |
| Energy contribution | -26.93 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
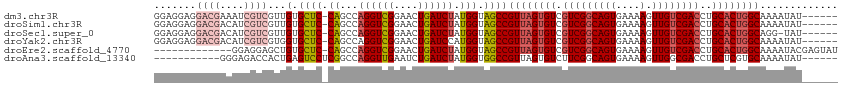
>dm3.chr3R 8065823 107 + 27905053 GGAGGAGGACGAAAUCGUCGUUGUGCUC-CAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUAU------ ((((.(.(((((.....))))).).)))-).(((((((((....)))))..))))....((((((((.(((((((((....).))))))))..)))))))).......------ ( -41.70, z-score = -3.28, R) >droSim1.chr3R 14194701 107 - 27517382 GGAGGAGGACGACAUCGUCGUUGUGCUC-CAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUAU------ ((((.(.((((((...)))))).).)))-).(((((((((....)))))..))))....((((((((.(((((((((....).))))))))..)))))))).......------ ( -43.50, z-score = -3.34, R) >droSec1.super_0 7212818 106 - 21120651 GGAGGAGGACGACAUCGUCGUUGUGCUC-CAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAGG-UAU------ ((((.(.((((((...)))))).).)))-).(((((((((....)))))..)))).(((((((((((.(((((((((....).))))))))..)))))))).))-)..------ ( -46.40, z-score = -3.59, R) >droYak2.chr3R 12382672 107 + 28832112 GGAGGAGGACGACAUCGUCGUGGUGCUC-CAGCCAGGUCGGAACUGAUCCAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUAU------ ((.((((.((.((......)).)).)))-).(((((((((....))).)).))))..))((((((((.(((((((((....).))))))))..)))))))).......------ ( -42.10, z-score = -2.52, R) >droEre2.scaffold_4770 4193097 100 - 17746568 -------------GGAGGAGCUGUGCUC-CAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUACGAGUAU -------------((.(((((...))))-).(((((((((....)))))..))))..))((((((((.(((((((((....).))))))))..))))))))............. ( -36.90, z-score = -2.27, R) >droAna3.scaffold_13340 19358785 97 - 23697760 -----------GGGAGACCACUGAGUCCUCGGCCAGGUUGAAUCUGAUCUAUGGUGGCCGUUAGUGUCUUCGGCAGUGAAAAGUUGGCGACCUGCUCGUGCAAAAUAU------ -----------((....))...((((....(..(((((...)))))..)...(((.(((((((.((.(....))).))).....)))).))).))))...........------ ( -27.10, z-score = 0.38, R) >consensus GGAGGAGGACGACAUCGUCGUUGUGCUC_CAGCCAGGUCGGAACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUAU______ .......((((....))))...(.(((....(((((((((....)))))..))))))))((((((((.((((((((.......))))))))..))))))))............. (-25.32 = -26.93 + 1.61)
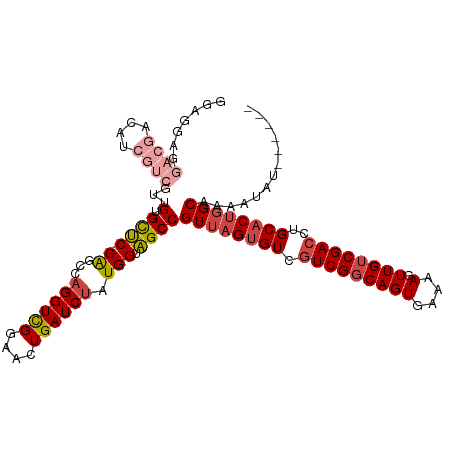
| Location | 8,065,863 – 8,065,964 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Shannon entropy | 0.43722 |
| G+C content | 0.42767 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -14.14 |
| Energy contribution | -15.23 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8065863 101 + 27905053 ---AACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAA------AUAU-AGUAAAUAGCGAAUGCA-AAAA-----AUAAAAGGCAGGC ---...............(((((((((((.(((((((((....).))))))))..))))))))...------....-........((....)).-....-----......))).... ( -27.20, z-score = -2.00, R) >droSim1.chr3R 14194741 101 - 27517382 ---AACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAA------AUAU-AGUAAAUAGCGAAUGCA-AAAA-----AUAAAAGGCAGGC ---...............(((((((((((.(((((((((....).))))))))..))))))))...------....-........((....)).-....-----......))).... ( -27.20, z-score = -2.00, R) >droSec1.super_0 7212858 100 - 21120651 ---AACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAG-------GUAU-AGUAAAUAGCGAAUGCA-AAAA-----AUAAAAGGCAGGC ---..(((.(((......(((((((((((.(((((((((....).))))))))..)))))))).)-------))..-........((....)).-....-----.....)))))).. ( -28.40, z-score = -1.92, R) >droYak2.chr3R 12382712 107 + 28832112 ---AACUGAUCCAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAA------AUAU-AGUAAAUAGAGAAUGCAAAAAAUAAGCAAAAAAGGCACGC ---............((.(((((((((((.(((((((((....).))))))))..))))))))...------....-.............(((.........))).....))))).. ( -27.90, z-score = -2.27, R) >droEre2.scaffold_4770 4193124 108 - 17746568 ---AACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAAAUACGAGUAU-AGUAAAUAGAGAAUGCGCAAAA-----AUAAAAGCCACGC ---..........((((....((((((((.(((((((((....).))))))))..)))))))).......(.((((-............)))).)....-----......))))... ( -25.10, z-score = -0.58, R) >droAna3.scaffold_13340 19358815 86 - 23697760 ---AUCUGAUCUAUGGUGGCCGUUAGUGUCUUCGGCAGUGAAAAGUUGGCGACCUGCUCGUGCAA------AAUAUUCACAAAAAGAAAAAACAC---------------------- ---......(((..(((.(((((((.((.(....))).))).....)))).)))(((....))).------.............)))........---------------------- ( -17.70, z-score = -0.16, R) >droWil1.scaffold_181089 12085097 96 - 12369635 AAACCCUGAUCUAUGGUAGCCGUUAGUGUCGCCGGCAGUGAAAAGUUGUCGACCUAAGCACAAAAUA----AAUAUCGACAAAUGUCUUGAACAAAAAAC----------------- .......(((.(((.((.((..((((.((((.((((........)))).))))))))))))...)))----...)))(((....))).............----------------- ( -15.90, z-score = -0.16, R) >consensus ___AACUGAUCUAUGGUAGCCGUUAGUGUCGUCGGCAGUGAAAAGUUGUCGACCUGCACUGGCAAA______AUAU_AGUAAAUAGCGAAUGCA_AAAA_____AUAAAAGGCAGGC .....................((((((((.((((((((.......))))))))..))))))))...................................................... (-14.14 = -15.23 + 1.08)

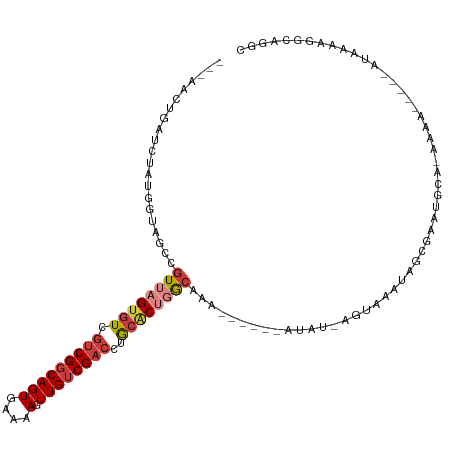
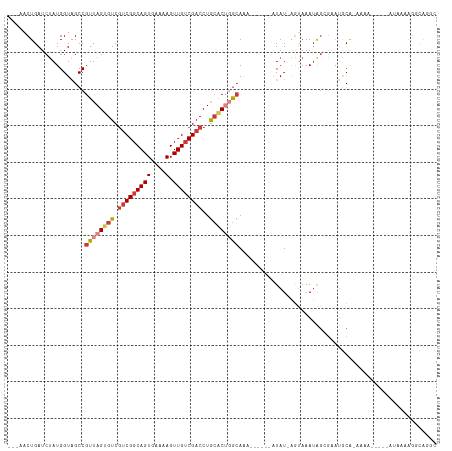
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:35 2011