
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,056,299 – 8,056,390 |
| Length | 91 |
| Max. P | 0.732201 |

| Location | 8,056,299 – 8,056,390 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.77 |
| Shannon entropy | 0.52625 |
| G+C content | 0.45109 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.89 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
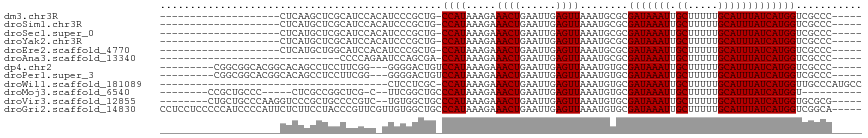
>dm3.chr3R 8056299 91 - 27905053 --------------------CUCAAGCUCGCAUCCACAUCCCGCUG-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- --------------------......................((.(-((((.....((((......))))........(((((((.((.....)))))))))))))).))..----- ( -17.10, z-score = -0.20, R) >droSim1.chr3R 14187071 91 + 27517382 --------------------CUCAUGCUCGCAUCCACAUCCCGCUG-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- --------------------......................((.(-((((.....((((......))))........(((((((.((.....)))))))))))))).))..----- ( -17.10, z-score = -0.13, R) >droSec1.super_0 7205196 91 + 21120651 --------------------CUCAUGCUCGCAUCCACAUCCCGCUG-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- --------------------......................((.(-((((.....((((......))))........(((((((.((.....)))))))))))))).))..----- ( -17.10, z-score = -0.13, R) >droYak2.chr3R 12373203 91 - 28832112 --------------------CUCAUGCUCGCAUCCACAUCCCGCUG-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- --------------------......................((.(-((((.....((((......))))........(((((((.((.....)))))))))))))).))..----- ( -17.10, z-score = -0.13, R) >droEre2.scaffold_4770 4183819 91 + 17746568 --------------------CUCAUGCUGGCAUCCACAUCCCGCUG-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- --------------------........(((..(((.........(-((((.....((((......))))..))).))(((((((.((.....))))))))).)))..))).----- ( -19.60, z-score = -0.48, R) >droAna3.scaffold_13340 19342367 81 + 23697760 ------------------------------CCCCAGAAUCCAGCGA-CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- ------------------------------............((((-((((.....((((......))))........(((((((.((.....)))))))))))))))))..----- ( -22.60, z-score = -3.87, R) >dp4.chr2 22835575 100 - 30794189 ---------CGGCGGCACGGCACAGCCUCCUUCGG---GGGGACUGUCCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- ---------.((((((((((.(((((((((....)---)))).)))))).......((((......))))....))))(((((((.((.....))))))))).....)))).----- ( -41.60, z-score = -4.59, R) >droPer1.super_3 5613474 100 - 7375914 ---------CGGCGGCACGGCACAGCCUCCUUCGG---GGGGACUGUCCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC----- ---------.((((((((((.(((((((((....)---)))).)))))).......((((......))))....))))(((((((.((.....))))))))).....)))).----- ( -41.60, z-score = -4.59, R) >droWil1.scaffold_181089 12081786 78 + 12369635 --------------------------------------CUCCUCGC-CCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUUGCCCAUGCC --------------------------------------......((-.(((.....((((......))))..))).))(((((((.((.....)))))))))((((....))))... ( -13.70, z-score = -0.66, R) >droMoj3.scaffold_6540 9557745 91 + 34148556 --------CCGCUGCCC-----CUCGCCGGCUCG-C--UUCGGCUGCCCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGU---------- --------.....(((.-----...((((((..(-(--....)).)))........((((......))))....).))(((((((.((.....)))))))))..)))---------- ( -21.20, z-score = -0.23, R) >droVir3.scaffold_12855 640772 102 + 10161210 --------CUGCUGCCCAAGGUCCCGCUGCCCCGUC--UGUGGCUGCCCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUGCGCG----- --------..((.(((((.(((.((((.........--.))))..)))((((....((((......))))...)))).(((((((.((.....))))))))).))).)))).----- ( -23.50, z-score = 0.10, R) >droGri2.scaffold_14830 1938883 112 - 6267026 CCUCCUCCCCCAUCCCCAUUCUCUUCCUACCCGUUCGUUGUGGCUGCCCAUAAAGAAACUGAAUUGAGUUAAAUGUGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGGCA----- ......((.((((...((((..(((........(((.((((((....)))))).)))........)))...))))...(((((((.((.....)))))))))))))..))..----- ( -20.29, z-score = -0.72, R) >consensus ____________________CUCAUGCUCGCACCCA_AUCCCGCUG_CCAUAAAGAAACUGAAUUGAGUUAAAUGCGCGAUAAAUUGCUUUUUGCAUUUAUCAUGGUCGCCC_____ ...............................................((((.....((((......))))........(((((((.((.....)))))))))))))........... (-11.89 = -11.89 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:33 2011