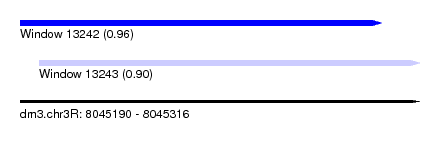
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,045,190 – 8,045,316 |
| Length | 126 |
| Max. P | 0.963029 |

| Location | 8,045,190 – 8,045,304 |
|---|---|
| Length | 114 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.65 |
| Shannon entropy | 0.41194 |
| G+C content | 0.53704 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8045190 114 + 27905053 ------ACCCGACAUGAGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCUCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCA ------....(((..(.((((((((.(((...((...((.....))...))...))).....)))))))))..)))......(((((((((((.........)))))))))))....... ( -38.00, z-score = -3.19, R) >droVir3.scaffold_12855 9498267 114 + 10161210 ------ACCCGACCAUCGUGCGCAACUCUUCGCGUUUGUUUUCCACCACCGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCCGUGCUGUUCGAACAGCGUGUCCACAUCUA ------....((..(((((((((........))........((((........))))..............))).)))).))((((((((.((.........)).))))))))....... ( -28.30, z-score = -0.69, R) >droGri2.scaffold_14830 1924280 114 + 6267026 ------ACCCGACCAUUGUGCGAAGUUCUUCGCGCUUGUUUUCGACCACCGUUUGGAUUUUCUUCUGCACCGCAUCGAUCUCGGACACGCCGUGCUGCUCGAACAGCGUGUCCACAUCUA ------...........(((((((....)))))))..(...(((((((.....))).........(((...)))))))...)((((((((.((.........)).))))))))....... ( -31.50, z-score = -0.94, R) >droMoj3.scaffold_6540 24568879 114 + 34148556 ------ACCCGACCAUGGUGCGCAGCUCUUCUCGCUUAUUCUCCACUACUGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUCUA ------....(((.(((.((.(((((.......))......((((........))))........))).)).))).).))..(((((((((((.........)))))))))))....... ( -33.50, z-score = -2.76, R) >droWil1.scaffold_181089 12070414 114 - 12369635 ------ACCCAACUUGUGUUCGCAGUUCCUCCCGCUUAUUCUCCACCACUGUCUGGAUCUUUUUGUGCACGGCAUCGAUUUCCGACACAUUAUUCUGUUCGAAUAAAGUGUCCACAUUUA ------........((((.(((..((.((...(((......((((........)))).......)))...)).))))).....(((((.((((((.....)))))).))))))))).... ( -20.72, z-score = -1.03, R) >droAna3.scaffold_13340 19331059 114 - 23697760 ------ACCCAACAUGAGUGCGAAGCUCCUCCCGCUUGUUCUCAACGAGUGACUGGAUCCGCUUGUGUAUCGCAUCGAUCUCGGACACACUGUGCUGCUCGAACAGCGUGUCCACGUUCA ------.....(((..((((.((...(((...(((((((.....)))))))...)))))))))..)))........((....((((((.((((.........)))).))))))....)). ( -37.10, z-score = -1.42, R) >droPer1.super_3 5601079 120 + 7375914 GCACUUACCCCACAUGAGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACAGUCUGUAUCUUUUUGUGCACUACAUCGAUCUCGGAUACGCUGUGCUGCUCAAACAGCGUAUCCACAUUUA .............(((((((((((((.....))(.((((........)))).)..........)))))))).))).......(((((((((((.........)))))))))))....... ( -33.80, z-score = -2.94, R) >dp4.chr2 22823454 120 + 30794189 GCACUUACCCCACAUGAGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACAGUCUGUAUCUUUUUGUGCACUACAUCGAUCUCGGAUACGCUGUGCUGCUCAAACAGCGUAUCCACAUUUA .............(((((((((((((.....))(.((((........)))).)..........)))))))).))).......(((((((((((.........)))))))))))....... ( -33.80, z-score = -2.94, R) >droSim1.chr3R 14175907 114 - 27517382 ------ACCCGACAUGGGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGUAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCA ------....(((...(((((((((......(((.(((...........))).)))......)))))))))..)))......(((((((((((.........)))))))))))....... ( -38.30, z-score = -2.77, R) >droSec1.super_0 7189291 114 - 21120651 ------ACCCGACAUGGGUGCGCAACUCCUCGCGCUUGUUUUCUACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCA ------....(((...(((((((((.(((...((...((.....))...))...))).....)))))))))..)))......(((((((((((.........)))))))))))....... ( -41.00, z-score = -3.36, R) >droYak2.chr3R 12361701 114 + 28832112 ------ACCCGACAUGGGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCA ------....(((...(((((((((.(((...((...((.....))...))...))).....)))))))))..)))......(((((((((((.........)))))))))))....... ( -40.30, z-score = -2.91, R) >droEre2.scaffold_4770 4171831 114 - 17746568 ------ACCCGACAUGGGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUUA ------....(((...((((((((((.....))........((((........))))......))))))))..)))......(((((((((((.........)))))))))))....... ( -41.80, z-score = -2.97, R) >anoGam1.chr2R 59731983 114 - 62725911 ------ACCCAACCAUCGUGCGAAGUUCCUCGCGCUUCAGCUCAAUCUCGUUCAGCACCCGCUUGUGCACCAAAUCGAUUUCGGCUACAUUGUGCUGCUUGAACAGUUGAUCCACAUCGA ------...........((((((......)))))).((((((.....(((..((((((..((....))......(((....))).......))))))..)))..)))))).......... ( -27.20, z-score = -0.73, R) >consensus ______ACCCGACAUGGGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACCGUCUGGAUCUUUUUGUGCACCGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUUUA .................(((((...........................................)))))............(((((((((((.........)))))))))))....... (-20.44 = -20.18 + -0.26)

| Location | 8,045,196 – 8,045,316 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Shannon entropy | 0.43365 |
| G+C content | 0.50256 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8045196 120 + 27905053 CAUGAGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCUCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUUAACAAAU .(..((((((........))))))..)..((((........))))....(((((..(((((((.......(((((((((((.........))))))))))))))...))))...))))). ( -38.31, z-score = -2.83, R) >droVir3.scaffold_12855 9498273 120 + 10161210 CCAUCGUGCGCAACUCUUCGCGUUUGUUUUCCACCACCGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCCGUGCUGUUCGAACAGCGUGUCCACAUCUAAUUGCAGUAAAU .....((((((........))........((((........)))).........)))).(((..(((...((((((((.((.........)).))))))))..)))....)))....... ( -30.50, z-score = -1.02, R) >droGri2.scaffold_14830 1924286 120 + 6267026 CCAUUGUGCGAAGUUCUUCGCGCUUGUUUUCGACCACCGUUUGGAUUUUCUUCUGCACCGCAUCGAUCUCGGACACGCCGUGCUGCUCGAACAGCGUGUCCACAUCUAAUUGCAGUAAAU .....(((((((....)))))))..........(((.....)))........(((((.......(....)((((((((.((.........)).)))))))).........)))))..... ( -32.70, z-score = -0.87, R) >droMoj3.scaffold_6540 24568885 120 + 34148556 CCAUGGUGCGCAGCUCUUCUCGCUUAUUCUCCACUACUGUUUGGAUUUUCUUCUGCACAGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUCUAAUUGCAAUAGAU .....((((..(((.......))).....((((........)))).........)))).(((..(((...(((((((((((.........)))))))))))..)))....)))....... ( -34.30, z-score = -2.33, R) >droWil1.scaffold_181089 12070420 120 - 12369635 CUUGUGUUCGCAGUUCCUCCCGCUUAUUCUCCACCACUGUCUGGAUCUUUUUGUGCACGGCAUCGAUUUCCGACACAUUAUUCUGUUCGAAUAAAGUGUCCACAUUUAAGUUUAACAAAC ..((((.(((..((.((...(((......((((........)))).......)))...)).))))).....(((((.((((((.....)))))).)))))))))................ ( -20.72, z-score = -0.40, R) >droAna3.scaffold_13340 19331065 120 - 23697760 CAUGAGUGCGAAGCUCCUCCCGCUUGUUCUCAACGAGUGACUGGAUCCGCUUGUGUAUCGCAUCGAUCUCGGACACACUGUGCUGCUCGAACAGCGUGUCCACGUUCAAGUUUAACAAAU ((..((((.((...(((...(((((((.....)))))))...)))))))))..)).........((....((((((.((((.........)))).))))))....))............. ( -35.00, z-score = -0.74, R) >droPer1.super_3 5601091 120 + 7375914 CAUGAGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACAGUCUGUAUCUUUUUGUGCACUACAUCGAUCUCGGAUACGCUGUGCUGCUCAAACAGCGUAUCCACAUUUAAAUUCAGUAGAU .(..((((((.((...))))))))..).......(((((...........)))))..((((...(....)(((((((((((.........))))))))))).............)))).. ( -33.90, z-score = -2.29, R) >dp4.chr2 22823466 120 + 30794189 CAUGAGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACAGUCUGUAUCUUUUUGUGCACUACAUCGAUCUCGGAUACGCUGUGCUGCUCAAACAGCGUAUCCACAUUUAAAUUCAGUAGAU .(..((((((.((...))))))))..).......(((((...........)))))..((((...(....)(((((((((((.........))))))))))).............)))).. ( -33.90, z-score = -2.29, R) >droSim1.chr3R 14175913 120 - 27517382 CAUGGGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGUAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUUAACAAAU .(..((((((........))))))..)........(((((((((((......)))))....)))......(((((((((((.........))))))))))).......)))......... ( -36.80, z-score = -1.97, R) >droSec1.super_0 7189297 120 - 21120651 CAUGGGUGCGCAACUCCUCGCGCUUGUUUUCUACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUUAACAAAU .((((((((((((.(((...((...((.....))...))...))).....))))))))).))).......(((((((((((.........)))))))))))................... ( -39.40, z-score = -2.45, R) >droYak2.chr3R 12361707 120 + 28832112 CAUGGGUGCGCAACUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUCAGGUUUAACAAAU .(..((((((........))))))..)..((((........))))....(((((..(((((((.......(((((((((((.........)))))))))))))))...)))...))))). ( -38.71, z-score = -2.08, R) >droEre2.scaffold_4770 4171837 120 - 17746568 CAUGGGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACCGACUGGAUCUUUUUGUGCACCGCGUCUAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACGUUUAGAUUUAACAAAU ....((((((((((.....))........((((........))))......))))))))..(((((....(((((((((((.........))))))))))).....)))))......... ( -41.90, z-score = -3.07, R) >anoGam1.chr2R 59731989 120 - 62725911 CCAUCGUGCGAAGUUCCUCGCGCUUCAGCUCAAUCUCGUUCAGCACCCGCUUGUGCACCAAAUCGAUUUCGGCUACAUUGUGCUGCUUGAACAGUUGAUCCACAUCGAUGUUCAGUAAAU .((((((((((......)))))).((((((.....(((..((((((..((....))......(((....))).......))))))..)))..))))))........)))).......... ( -31.10, z-score = -1.23, R) >consensus CAUGGGUGCGCAGCUCCUCGCGCUUGUUCUCCACCACCGUCUGGAUCUUUUUGUGCACCGCAUCGAUCUCGGACACGCUGUGCUGCUCAAACAGCGUGUCCACAUUUAAGUUUAACAAAU .....(((((...........................................)))))............(((((((((((.........)))))))))))................... (-20.44 = -20.18 + -0.26)
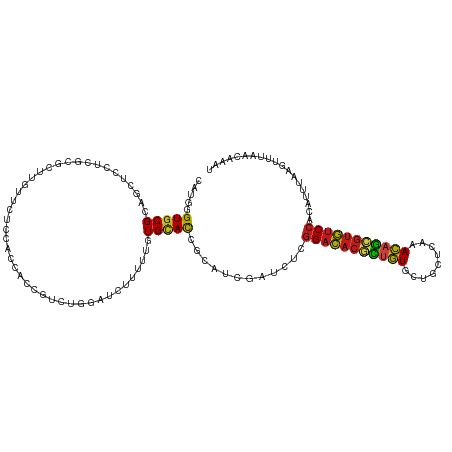
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:32 2011