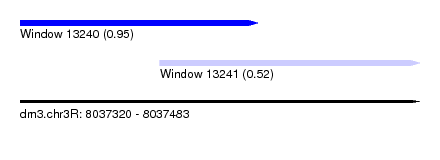
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,037,320 – 8,037,483 |
| Length | 163 |
| Max. P | 0.952334 |

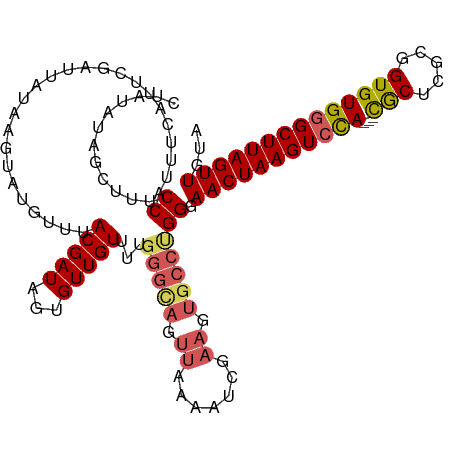
| Location | 8,037,320 – 8,037,417 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Shannon entropy | 0.32481 |
| G+C content | 0.40447 |
| Mean single sequence MFE | -33.96 |
| Consensus MFE | -19.31 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8037320 97 + 27905053 CUAUAUAGCUUUCCAUUUCAUUCGAGUAUAAGUAUGUUUACGAUAGUGUUGU----------------------GCCUGG-AACUAAGUCCACGACGCUCGCAGUGUGGGCUUAGUUGUA ........((...(((..((((...(((..........)))...))))..))----------------------)...))-(((((((((((((..(....)..)))))))))))))... ( -24.00, z-score = -0.62, R) >droEre2.scaffold_4770 4162443 116 - 17746568 -CUUAUAGCUUUCCAUUUCAUUCGAUUAUAAGUAUGCUUACGAUAAUGUUGUUUUGGGUAGUUAAAAUCGAAGUGCCUGGGAACUAAGUCCA---UACUUGCUGUGUGAGCUUAGUUGUA -..........((((.....(((((((.(((.(((.(..(((((...)))))...).))).))).))))))).....))))(((((((..((---(((.....)))))..)))))))... ( -33.00, z-score = -3.13, R) >droYak2.chr3R 12354099 115 + 28832112 --CUUUAGCUUUCCAUUUCAUUCAUCGAUAAGUAGGCUUACGAUAAUGUUGUUUUGGGUAGUUAAAAGCGAAGUGCCCGGGAACUAAGUCUA---UACUUGCUGUGUGGGCUUAGUUGUA --.........(((((..((((.((((.((((....))))))))))))..))...(((((.((.......)).))))))))(((((((((((---(((.....))))))))))))))... ( -36.50, z-score = -3.19, R) >droSec1.super_0 7181509 117 - 21120651 CUAUAUAGCUUUCCAUUUCAUUCGAUUAUAAGUAUGUUUACGAUAGUGUUGUUUUGGGCAGUUAAGAUCGAAGUGCCUGGGAACUAAGUCCA---CGCUCGCGGUGUGGGCUUAGUUGUA ...........((((.....(((((((.(((...((((((.((((....)))).)))))).))).))))))).....))))(((((((((((---(((.....))))))))))))))... ( -37.20, z-score = -2.94, R) >droSim1.chr3R 14168354 117 - 27517382 CUAUAUAGCUUUCCAUUUCAUUCGAUUAUAACUAUGUUUACGAUAGUGUUGUUUUGGGCAGUUAAGAUCGAAGUGCCUGGGAACUAAGUCCA---CGCUCGCGGUGUGGGCUUAGUUGUA ...........((((.....(((((((.(((((..(((((.((((....)))).)))))))))).))))))).....))))(((((((((((---(((.....))))))))))))))... ( -39.10, z-score = -3.49, R) >consensus CUAUAUAGCUUUCCAUUUCAUUCGAUUAUAAGUAUGUUUACGAUAGUGUUGUUUUGGGCAGUUAAAAUCGAAGUGCCUGGGAACUAAGUCCA___CGCUCGCGGUGUGGGCUUAGUUGUA ...........(((........((((.....(((....)))......))))....(((((.((.......)).))))))))(((((((((((...(((.....))))))))))))))... (-19.31 = -20.06 + 0.75)

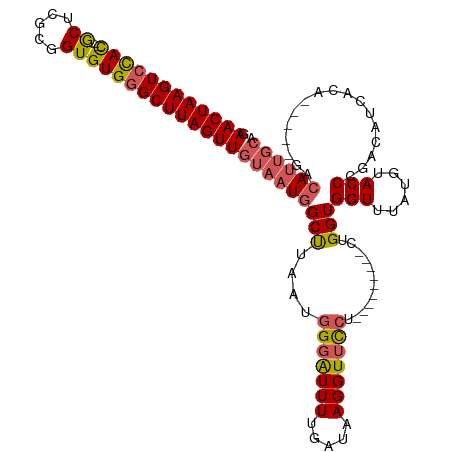
| Location | 8,037,377 – 8,037,483 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Shannon entropy | 0.24565 |
| G+C content | 0.46062 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -22.46 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8037377 106 + 27905053 GAACUAAGUCCACGACGCUCGCAGUGUGGGCUUAGUUGUAAUGGCUUAAUGGGAUUUUGAUAAGGUUCCU---------CUGGUGGUUUAUGUACCCGACAUCACA-----GACAUUGCA .(((((((((((((..(....)..)))))))))))))((((((.......(((((((.....)))))))(---------((((.(((......))))(....).))-----)))))))). ( -34.30, z-score = -1.98, R) >droEre2.scaffold_4770 4162522 112 - 17746568 GAACUAAGUCCAU---ACUUGCUGUGUGAGCUUAGUUGUAAUGGCCUAAUGGGGUUUUGAUAAGGUUCCUCUGGUUCCUCUGGUGGUUUGUGUACCCGACAUCACA-----GAAAUUGCA .(((((((..(((---((.....)))))..)))))))(((((((((....((..(((.....)))..))...))))..((((.(((((((......))).))))))-----)).))))). ( -31.10, z-score = -0.72, R) >droYak2.chr3R 12354177 108 + 28832112 GAACUAAGUCUAU---ACUUGCUGUGUGGGCUUAGUUGUAAUGGCUUAAUGGGGUUUUGAUAAGGUUCCU---------CUGGUGGUUUAUGUACCCGACAUCACUCACCCGACAUUGCA .((((((((((((---((.....))))))))))))))((((((.......((..(((.....)))..))(---------(.((((..(.((((.....)))).)..)))).)))))))). ( -36.40, z-score = -2.67, R) >droSec1.super_0 7181589 97 - 21120651 GAACUAAGUCCAC---GCUCGCGGUGUGGGCUUAGUUGUAAUGGCUUAAUGGGAUUUCGAUAAGGUGUA----------CUGGUGGUUUAUGUACCCGACAUCACA-----GACA----- .((((((((((((---((.....))))))))))))))..........................(((((.----------(.((((.......)))).))))))...-----....----- ( -30.00, z-score = -1.25, R) >droSim1.chr3R 14168434 103 - 27517382 GAACUAAGUCCAC---GCUCGCGGUGUGGGCUUAGUUGUAAUGGCUUAAUGGGAUUUCGAUAAGGUUCCU---------CUGGUGGUUUAUGUACCCGACAUCACA-----GACAUUGCA .((((((((((((---((.....))))))))))))))((((((.......(((((((.....)))))))(---------((((.(((......))))(....).))-----)))))))). ( -37.20, z-score = -2.98, R) >consensus GAACUAAGUCCAC___GCUCGCGGUGUGGGCUUAGUUGUAAUGGCUUAAUGGGAUUUUGAUAAGGUUCCU_________CUGGUGGUUUAUGUACCCGACAUCACA_____GACAUUGCA .((((((((((((...((.....))))))))))))))((((((.......(((((((.....)))))))...........(((((..............))))).........)))))). (-22.46 = -23.02 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:30 2011