
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 8,029,524 – 8,029,655 |
| Length | 131 |
| Max. P | 0.927720 |

| Location | 8,029,524 – 8,029,616 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.12 |
| Shannon entropy | 0.56446 |
| G+C content | 0.33179 |
| Mean single sequence MFE | -15.86 |
| Consensus MFE | -5.41 |
| Energy contribution | -5.66 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
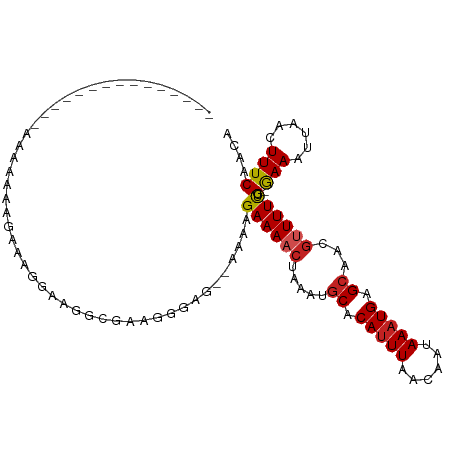
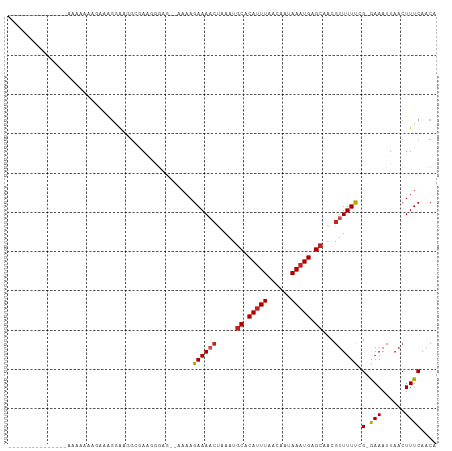
>dm3.chr3R 8029524 92 + 27905053 --------------UAAAAAAAUGAGGGGUGGUGAAGGAAG--GAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACA --------------........((((((.((((....((((--((.((....))...(((.((((((....)))))).)))...)))))).-...)))).))))))... ( -14.60, z-score = -1.71, R) >droSim1.chr3R 14155617 93 - 27517382 -------------UGAAAAAAAUGUAGGGAGGCGAAGGGCG--GAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACA -------------.........(((.(..((.(((((((((--...((....))...(((.((((((....)))))).))).)))))))))-........))..).))) ( -18.50, z-score = -2.86, R) >droSec1.super_0 7173806 93 - 21120651 -------------UGAAAAAAAUGUAGGGAGGCGAAGGGCG--GAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACA -------------.........(((.(..((.(((((((((--...((....))...(((.((((((....)))))).))).)))))))))-........))..).))) ( -18.50, z-score = -2.86, R) >droYak2.chr3R 12345744 89 + 28832112 -----------------UGAAAAAAGGGAUGGCAAACAGAC--GAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACA -----------------.........(..((.....))..)--(((((.........(((.((((((....)))))).))).((.....))-........))))).... ( -13.80, z-score = -2.12, R) >droEre2.scaffold_4770 4154752 98 - 17746568 --------UAAAAAAGAGGAACAAGGGGAAGGCGAACAGAG--GAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACA --------..................((((((((.....((--.........))...(((.((((((....)))))).))).))))))))(-(((......)))).... ( -14.60, z-score = -2.35, R) >droAna3.scaffold_13340 19315724 78 - 23697760 ---------------------------CGAAAUGAAAAUAG--CAAAGAAAACUAAAUGCGCAUUUAACAAUAAAUGAGAAACGUUUUC-G-GAAAUUAACUUUCAACA ---------------------------.((((.(..(((..--(...((((((......(.((((((....)))))).)....))))))-.-)..)))..))))).... ( -9.20, z-score = -0.76, R) >dp4.chr2 22807359 85 + 30794189 -------------------AGAGAAGGAAGUGCGAA-GGAG--AAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUU-G-GAAAUUAACUUUCAACA -------------------......((((((.(...-.)..--....((((((....(((.((((((....)))))).)))..))))))-.-.......)))))).... ( -14.70, z-score = -2.16, R) >droPer1.super_3 5584995 85 + 7375914 -------------------AAAGAAGGAAGUGCGAA-GGAG--AAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUU-G-GAAAUUAACUUUCAACA -------------------......((((((.(...-.)..--....((((((....(((.((((((....)))))).)))..))))))-.-.......)))))).... ( -14.70, z-score = -2.25, R) >droVir3.scaffold_12855 9480937 107 + 10161210 GGGGAAAUGAAAUGAAAAAAAAGAAAGAAAGGAAAAUGGCAGCAAAAGAAAACCAAAUGCGCAUUUAGCAG-AAAUGAGCUGCGUUUUCCG-AAAAUUAACUUUCAACA ......................(((((...((((((((.((((....(.....)...(((.......))).-......)))))))))))).-........))))).... ( -19.50, z-score = -1.19, R) >droMoj3.scaffold_6540 24553808 100 + 34148556 -------CCAAAGAAAAAUGAAGGGAACAAGGAAAAUGGCAGCAAAAGAAAUCCAAAUGCGCACUUAGCAG-AAAUGAGCUGCGUUUUCAG-AAAAUUAACUUUCAACA -------...........((((((.......(((((((.((((....(.....)...(((.......))).-......)))))))))))..-........))))))... ( -18.93, z-score = -0.97, R) >droGri2.scaffold_14830 1909967 91 + 6267026 ---------AAUUGGAGAGAAAGAGAGAA--GGAAGUGGCAUCAAAAGAAA------UGCGCAUUUAGCAG-AAAUGAGCUGCGUUUUUGGGAAAAUUAACUUUCAACA ---------.............(((((..--..(((((((((........)------))).)))))..(((-(((((.....))))))))..........))))).... ( -17.40, z-score = -0.68, R) >consensus _______________AAAAAAAGAAAGGAAGGCGAAGGGAG__AAAAGAAAACUAAAUGCACAUUUAACAAUAAAUGAGCAACGUUUUUCG_GAAAUUAACUUUCAACA ...............................................((((((.....((.(((((......))))).))...)))))).................... ( -5.41 = -5.66 + 0.26)

| Location | 8,029,524 – 8,029,616 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 72.12 |
| Shannon entropy | 0.56446 |
| G+C content | 0.33179 |
| Mean single sequence MFE | -13.51 |
| Consensus MFE | -5.09 |
| Energy contribution | -5.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8029524 92 - 27905053 UGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC--CUUCCUUCACCACCCCUCAUUUUUUUA-------------- ....((((.....))))-.(((((....(((.((((((....)))))).))).....)))))....--...........................-------------- ( -12.20, z-score = -2.49, R) >droSim1.chr3R 14155617 93 + 27517382 UGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC--CGCCCUUCGCCUCCCUACAUUUUUUUCA------------- ...((((((........-.(((((....(((.((((((....)))))).))).....)))))....--.((.....))............))))))------------- ( -13.80, z-score = -2.45, R) >droSec1.super_0 7173806 93 + 21120651 UGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC--CGCCCUUCGCCUCCCUACAUUUUUUUCA------------- ...((((((........-.(((((....(((.((((((....)))))).))).....)))))....--.((.....))............))))))------------- ( -13.80, z-score = -2.45, R) >droYak2.chr3R 12345744 89 - 28832112 UGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC--GUCUGUUUGCCAUCCCUUUUUUCA----------------- ....((..((.(((...-((((((((..(((.((((((....)))))).)))....)))...))))--)...))).))..))..........----------------- ( -13.60, z-score = -2.08, R) >droEre2.scaffold_4770 4154752 98 + 17746568 UGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC--CUCUGUUCGCCUUCCCCUUGUUCCUCUUUUUUA-------- ....(((.((.(((...-.(((((((..(((.((((((....)))))).)))....)))...))))--....))).)).)))...................-------- ( -12.60, z-score = -2.65, R) >droAna3.scaffold_13340 19315724 78 + 23697760 UGUUGAAAGUUAAUUUC-C-GAAAACGUUUCUCAUUUAUUGUUAAAUGCGCAUUUAGUUUUCUUUG--CUAUUUUCAUUUCG--------------------------- ...(((((((.......-.-((((((....(.((((((....)))))).)......))))))....--..))))))).....--------------------------- ( -11.44, z-score = -1.34, R) >dp4.chr2 22807359 85 - 30794189 UGUUGAAAGUUAAUUUC-C-AAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUU--CUCC-UUCGCACUUCCUUCUCU------------------- ....((((.....))))-.-.(((((..(((.((((((....)))))).)))....))))).....--....-.................------------------- ( -11.00, z-score = -2.08, R) >droPer1.super_3 5584995 85 - 7375914 UGUUGAAAGUUAAUUUC-C-AAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUU--CUCC-UUCGCACUUCCUUCUUU------------------- ....((((.....))))-.-.(((((..(((.((((((....)))))).)))....))))).....--....-.................------------------- ( -11.00, z-score = -1.93, R) >droVir3.scaffold_12855 9480937 107 - 10161210 UGUUGAAAGUUAAUUUU-CGGAAAACGCAGCUCAUUU-CUGCUAAAUGCGCAUUUGGUUUUCUUUUGCUGCCAUUUUCCUUUCUUUCUUUUUUUUCAUUUCAUUUCCCC ....(((((....))))-)((((((.(((((......-..(((((((....)))))))........)))))..)))))).............................. ( -20.89, z-score = -2.69, R) >droMoj3.scaffold_6540 24553808 100 - 34148556 UGUUGAAAGUUAAUUUU-CUGAAAACGCAGCUCAUUU-CUGCUAAGUGCGCAUUUGGAUUUCUUUUGCUGCCAUUUUCCUUGUUCCCUUCAUUUUUCUUUGG------- ...((((.(..(((...-..(((((.(((((......-...((((((....)))))).........)))))..)))))...)))..)))))...........------- ( -15.77, z-score = 0.09, R) >droGri2.scaffold_14830 1909967 91 - 6267026 UGUUGAAAGUUAAUUUUCCCAAAAACGCAGCUCAUUU-CUGCUAAAUGCGCA------UUUCUUUUGAUGCCACUUCC--UUCUCUCUUUCUCUCCAAUU--------- ....(((((.................((((.......-))))....((.(((------((......))))))).....--......))))).........--------- ( -12.50, z-score = -1.64, R) >consensus UGUUGAAAGUUAAUUUC_CGAAAAACGUUGCUCAUUUAUUGUUAAAUGUGCAUUUAGUUUUCUUUC__CUCCAUUCGCCUUCCCUUCUUUUUUU_______________ ....((((((((((((....)))......((.(((((......))))).))..)))))))))............................................... ( -5.09 = -5.32 + 0.22)

| Location | 8,029,547 – 8,029,655 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.76 |
| Shannon entropy | 0.33507 |
| G+C content | 0.34135 |
| Mean single sequence MFE | -22.79 |
| Consensus MFE | -12.26 |
| Energy contribution | -12.37 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
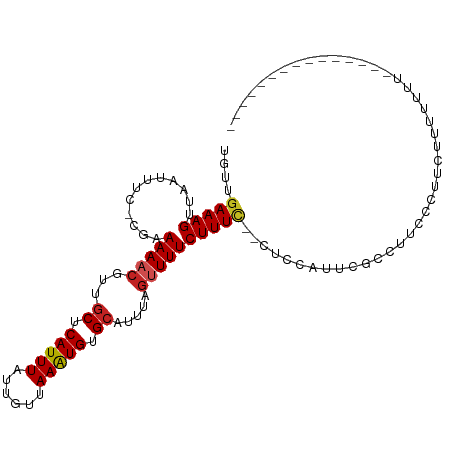
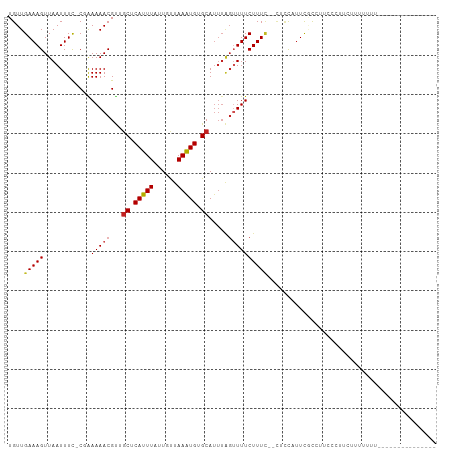
>dm3.chr3R 8029547 108 + 27905053 --------GAAGGAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCACCCAUAACG --------...((...((((((....(((.((((((....)--))))).)))..))))))((-(((..((((((((((((.....))))))))-.)...)))..)))))...))...... ( -21.50, z-score = -2.70, R) >droSim1.chr3R 14155641 108 - 27517382 --------GGCGGAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCACCCAUAACG --------.((((((.((....((((((((((((((....)--)))))..............-.........((((((((.....))))))))-)))))))))))))))).......... ( -26.70, z-score = -3.95, R) >droSec1.super_0 7173830 108 - 21120651 --------GGCGGAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCACCCAUAACG --------.((((((.((....((((((((((((((....)--)))))..............-.........((((((((.....))))))))-)))))))))))))))).......... ( -26.70, z-score = -3.95, R) >droYak2.chr3R 12345764 107 + 28832112 --------AGACGAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCACCCA-AACG --------...(....).........(((.((((((....)--))))).))).(((((..((-(((..((((((((((((.....))))))))-.)...)))..))))).....)-)))) ( -20.90, z-score = -2.68, R) >droEre2.scaffold_4770 4154781 107 - 17746568 --------AGAGGAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUUCG-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCACCGA-AACG --------...((...((((((....(((.((((((....)--))))).)))..))))))((-(((..((((((((((((.....))))))))-.)...)))..)))))..))..-.... ( -21.70, z-score = -2.34, R) >droAna3.scaffold_13340 19315734 107 - 23697760 --------AUAGCAAAGAAAACUAAAUGCGCAUUUAACAAU--AAAUGAGAAACGUUUUC-G-GAAAUUAACUUUCAACAACAUGUGUUGAAA-CGCAUUUAUCUUCCGCCCUUGCAACG --------...((((.((((((......(.((((((....)--))))).)....))))))-(-(((..((((((((((((.....))))))))-.)...)))..))))....)))).... ( -22.10, z-score = -2.53, R) >dp4.chr2 22807376 107 + 30794189 --------GGAGAAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUU-G-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCUCUCAUAACG --------.((((...((((((....(((.((((((....)--))))).)))..))))))-(-(((..((((((((((((.....))))))))-.)...)))..))))..))))...... ( -23.30, z-score = -3.29, R) >droPer1.super_3 5585012 107 + 7375914 --------GGAGAAAAGAAAACUAAAUGCACAUUUAACAAU--AAAUGAGCAACGUUUUU-G-GAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGCUCUCAUAACG --------.((((...((((((....(((.((((((....)--))))).)))..))))))-(-(((..((((((((((((.....))))))))-.)...)))..))))..))))...... ( -23.30, z-score = -3.29, R) >droWil1.scaffold_181089 12026301 109 - 12369635 --------AAAGAAAUUAAACCUAAAUGCACAUUUAGAAGAGAAAAUGAGCAGCGUUUUG---GAAAUUAACUUUCAACAACAUGUGUUGAAAAUGCAUUUAUCUUUUAGGGUUUUAACA --------............(((((((((.(((((........))))).)))(((((((.---((((.....))))..((((....)))))))))))........))))))......... ( -22.00, z-score = -1.75, R) >droVir3.scaffold_12855 9480968 115 + 10161210 GAAAAUGGCAGCAAAAGAAAACCAAAUGCGCAUUUAGCAG---AAAUGAGCUGCGUUUUCCG-AAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGGACUCAUAACG (((.((((..(((...((((((((...((.(((((.....---))))).)))).))))))..-.........((((((((.....))))))))-)))..)))).)))............. ( -20.30, z-score = 0.07, R) >droMoj3.scaffold_6540 24553832 115 + 34148556 GAAAAUGGCAGCAAAAGAAAUCCAAAUGCGCACUUAGCAG---AAAUGAGCUGCGUUUUCAG-AAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCCGGACUCAUAACG (((((((.((((....(.....)...(((.......))).---......)))))))))))((-(.((.....((((((((.....))))))))-.....)).)))............... ( -20.90, z-score = -0.04, R) >droGri2.scaffold_14830 1909987 110 + 6267026 GGAAGUGGCAUCAAAAGAAAU------GCGCAUUUAGCAG---AAAUGAGCUGCGUUUUUGGGAAAAUUAACUUUCAACAACAUGUGUUGAAA-UGCAUUUAUCUUCUGGACUCAUAACG ..(((((((((........))------)).)))))..(((---(((((((.((((((((((..(........)..)))((((....)))))))-))))))))).)))))........... ( -24.10, z-score = -0.71, R) >consensus ________GGAGAAAAGAAAACUAAAUGCACAUUUAACAAU__AAAUGAGCAACGUUUUUCG_GAAAUUAACUUUCAACAACAUGUGUUGAAA_UGCAUUUAUCUUCCGCACUCAUAACG .......................((((((((((((........)))))........................((((((((.....)))))))).)))))))................... (-12.26 = -12.37 + 0.12)

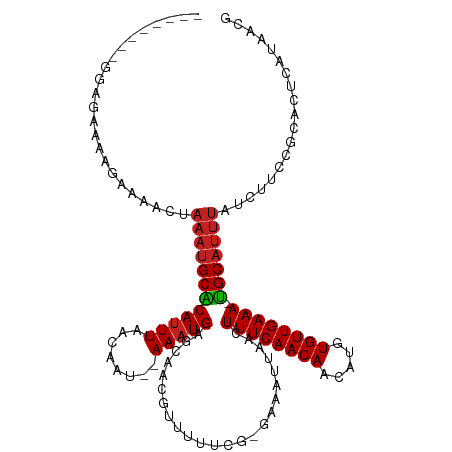

| Location | 8,029,547 – 8,029,655 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Shannon entropy | 0.33507 |
| G+C content | 0.34135 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.96 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 8029547 108 - 27905053 CGUUAUGGGUGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUCCUUC-------- ......(((..((((((.(((.....-((((((((.....)))))))).))).))))-)).(((((..(((.(((((--(....)))))).)))....)))))...)))...-------- ( -26.50, z-score = -2.56, R) >droSim1.chr3R 14155641 108 + 27517382 CGUUAUGGGUGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUCCGCC-------- ..........((((((((.....((.-((((((((.....)))))))))).....))-.(((((....(((.(((((--(....)))))).))).....))))).)))))).-------- ( -28.60, z-score = -2.76, R) >droSec1.super_0 7173830 108 + 21120651 CGUUAUGGGUGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUCCGCC-------- ..........((((((((.....((.-((((((((.....)))))))))).....))-.(((((....(((.(((((--(....)))))).))).....))))).)))))).-------- ( -28.60, z-score = -2.76, R) >droYak2.chr3R 12345764 107 - 28832112 CGUU-UGGGUGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUCGUCU-------- ((((-(.....((((((.(((.....-((((((((.....)))))))).))).))))-))..))))).(((.(((((--(....)))))).)))..................-------- ( -25.70, z-score = -2.01, R) >droEre2.scaffold_4770 4154781 107 + 17746568 CGUU-UCGGUGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-CGAAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUCCUCU-------- ....-..((..((((((.(((.....-((((((((.....)))))))).))).))))-)).(((((..(((.(((((--(....)))))).)))....)))))....))...-------- ( -25.70, z-score = -2.52, R) >droAna3.scaffold_13340 19315734 107 + 23697760 CGUUGCAAGGGCGGAAGAUAAAUGCG-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-C-GAAAACGUUUCUCAUUU--AUUGUUAAAUGCGCAUUUAGUUUUCUUUGCUAU-------- ....((((((((....).((((((((-((((((((.....)))))))).........-.-............(((((--(....))))))))))))))....)))))))...-------- ( -26.10, z-score = -1.80, R) >dp4.chr2 22807376 107 - 30794189 CGUUAUGAGAGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-C-AAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUUCUCC-------- ......(((((.((((((((((((((-((((((((.....)))))))).........-.-............(((((--(....))))))))))))).))))))).))))).-------- ( -27.90, z-score = -3.53, R) >droPer1.super_3 5585012 107 - 7375914 CGUUAUGAGAGCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUC-C-AAAAACGUUGCUCAUUU--AUUGUUAAAUGUGCAUUUAGUUUUCUUUUCUCC-------- ......(((((.((((((((((((((-((((((((.....)))))))).........-.-............(((((--(....))))))))))))).))))))).))))).-------- ( -27.90, z-score = -3.53, R) >droWil1.scaffold_181089 12026301 109 + 12369635 UGUUAAAACCCUAAAAGAUAAAUGCAUUUUCAACACAUGUUGUUGAAAGUUAAUUUC---CAAAACGCUGCUCAUUUUCUCUUCUAAAUGUGCAUUUAGGUUUAAUUUCUUU-------- .((((((..((((((....((((..((((((((((.....))))))))))..)))).---........(((.(((((........))))).)))))))))))))))......-------- ( -20.70, z-score = -2.58, R) >droVir3.scaffold_12855 9480968 115 - 10161210 CGUUAUGAGUCCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUU-CGGAAAACGCAGCUCAUUU---CUGCUAAAUGCGCAUUUGGUUUUCUUUUGCUGCCAUUUUC .(((.....((((((((.(((.....-((((((((.....)))))))).))).))))-)))).)))(((((......---..(((((((....)))))))........)))))....... ( -30.49, z-score = -2.16, R) >droMoj3.scaffold_6540 24553832 115 - 34148556 CGUUAUGAGUCCGGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUU-CUGAAAACGCAGCUCAUUU---CUGCUAAGUGCGCAUUUGGAUUUCUUUUGCUGCCAUUUUC ...........((((((((....((.-((((((((.....))))))))))..)))))-))).....(((((......---...((((((....)))))).........)))))....... ( -26.77, z-score = -0.85, R) >droGri2.scaffold_14830 1909987 110 - 6267026 CGUUAUGAGUCCAGAAGAUAAAUGCA-UUUCAACACAUGUUGUUGAAAGUUAAUUUUCCCAAAAACGCAGCUCAUUU---CUGCUAAAUGCGC------AUUUCUUUUGAUGCCACUUCC ...........(((((((..(((((.-((((((((.....))))))))..................((((.......---)))).......))------))))))))))........... ( -22.30, z-score = -1.44, R) >consensus CGUUAUGAGUGCGGAAGAUAAAUGCA_UUUCAACACAUGUUGUUGAAAGUUAAUUUC_CGAAAAACGUUGCUCAUUU__AUUGUUAAAUGUGCAUUUAGUUUUCUUUUCUCC________ ............(((((((((((((..((((((((.....))))))))........................(((((........))))).)))))))....))))))............ (-15.90 = -15.96 + 0.06)


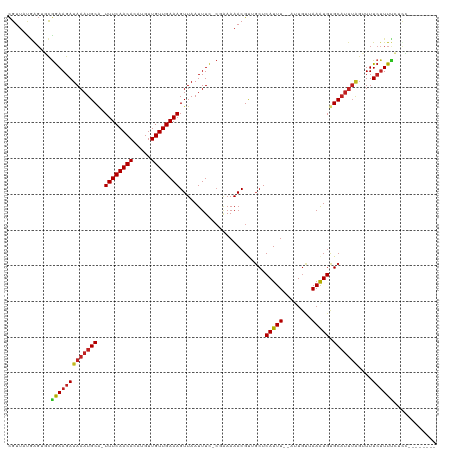
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:28 2011