
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,957,023 – 7,957,165 |
| Length | 142 |
| Max. P | 0.998931 |

| Location | 7,957,023 – 7,957,165 |
|---|---|
| Length | 142 |
| Sequences | 11 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 67.76 |
| Shannon entropy | 0.70249 |
| G+C content | 0.49468 |
| Mean single sequence MFE | -44.43 |
| Consensus MFE | -32.85 |
| Energy contribution | -31.85 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
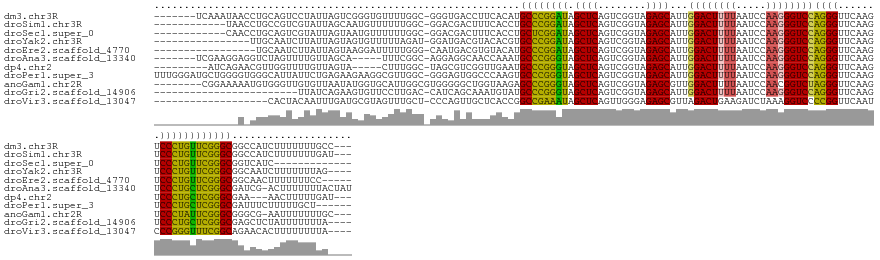
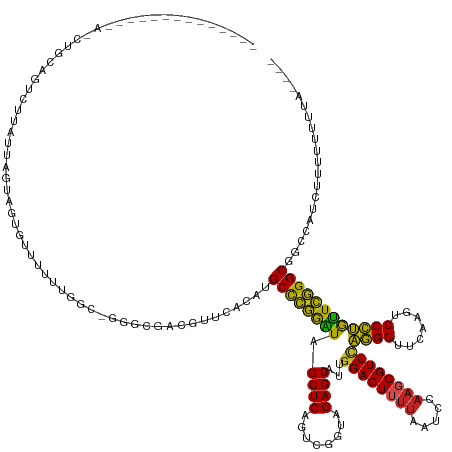
>dm3.chr3R 7957023 142 + 27905053 -------UCAAAUAACCUGCAGUCCUAUUAGUCGGGUGUUUUGGC-GGGUGACCUUCACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCCAUCUUUUUUUGCC--- -------.((((..((((((..........).)))))....((((-((....))......((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))))))......))))..--- ( -48.60, z-score = -1.64, R) >droSim1.chr3R 14082390 137 - 27517382 ------------UAACCUGCCGUCGUAUUAGCAAUGUUUUUUGGC-GGACGACUUUCACCUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCCAUCUUUUUUUGAU--- ------------......((((((((....((((......)).))-..)))))........(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))))..............--- ( -45.70, z-score = -1.76, R) >droSec1.super_0 7101578 127 - 21120651 ------------CAACCUGCAGUCGUAUUAGUAAUGUUUUUUGGC-GGACGACUUUCACCUGCUCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGUCAUC------------- ------------........((((((....((...........))-..)))))).....(((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))).....------------- ( -39.20, z-score = -1.29, R) >droYak2.chr3R 7764400 132 + 28832112 ----------------UUGCAAUCUUAUUAGUAGUGUUUUUAGAU-GGAUGACGUACACGUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAAUCUUUUUUUAG---- ----------------((((.............((((..(((...-...)))...)))).((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))))))...........---- ( -40.50, z-score = -2.07, R) >droEre2.scaffold_4770 14848456 130 - 17746568 -----------------UGCAAUCUUAUUAGUAAGGAUUUUUGGG-CAAUGACGUGUACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCAACUUUUUUUCC----- -----------------(((((((((.......))))))..((..-((......))..))((((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))))))...........----- ( -39.00, z-score = -1.25, R) >droAna3.scaffold_13340 5404152 139 + 23697760 -------UCGAAGGAGGUCUAGUUUUGUUAGCA-----UUUCGGC-AGGAGGCAACCAAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCG-ACUUUUUUUACUAU -------..((((((((((..((((.(((.((.-----((((...-.))))))))).))))(((((((((((((........))))..(((((((((.....)))))))))(((.......))))))))))))....)-)))))))))..... ( -50.20, z-score = -2.71, R) >dp4.chr2 19721640 132 + 30794189 ---------AUCAGAACGUUGGUUUUGUUAGUA-----CUUUGGC-UAGCGUCGGUUGAAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAA---AACUUUUUGAU--- ---------(((((((((((((((..((....)-----)...)))-)))))).((((...((((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))..---)))).))))))--- ( -49.80, z-score = -3.39, R) >droPer1.super_3 2464834 146 + 7375914 UUUGGGAUGCUGGGGUGGGCAUUAUUCUGAGAAGAAGGCGUUGGC-GGGAGUGGCCCAAGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUUUCUUUUUGCU------ ...(..(((((.(((..((.....((((....))))(((.(((((-..((((.((((........)))).)))).)))))..((((.((((((((((.....)))))))))).))))..)))))..))).))).))..)........------ ( -53.90, z-score = -0.43, R) >anoGam1.chr2R 51278841 141 + 62725911 --------CGGAAAAAUGUGGGUUGUGUUAAUAUGGUGCAUUGGCGUGGGGGCUGGUAAGAGCCCGGGUAGCUCAGUCGGUAGAGCGUUGGACUUUUAAUCCAACGGUCUAGGGUUCAAGUCCCUAUUCGGGCGGGCG-AAUUUUUUUGC--- --------(((((((((.(.(.((((((((((.......))))))(((((((((.....(((((((((...)))......((((.(((((((.......))))))).)))))))))).)))))))))....)))).).-)))))))))).--- ( -47.50, z-score = -1.65, R) >droGri2.scaffold_14906 11425882 124 + 14172833 ------------------------UUAUCAGAAGUGUUCCUUGAC-CAUCAGCAAAUGUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCUCUAUUUUUUUA---- ------------------------........((.((((.(((..-...))).........(((((((((((((........))))..(((((((((.....)))))))))(((.......)))))))))))))))).)).........---- ( -39.40, z-score = -1.83, R) >droVir3.scaffold_13047 1772746 129 + 19223366 -------------------CACUACAAUUUGAUGCGUAGUUUGCU-CCCAGUUGCUCACCGGCCGAAAUAGCUCAGUUGGGAGAGCGUUAGACUGAAGAUCUAAAGGUCCCCGGUUCAAUCCCGGGUUUCGGCAGAACACUUUUUUUUA---- -------------------..((((..........))))((((((-..(((((((((.(((((.((......)).)))))..))))....)))))..........((..(((((.......)))))..)))))))).............---- ( -34.90, z-score = -0.30, R) >consensus ______________A_CUGCAGUCUUAUUAGUAGUGUUUUUUGGC_GGGCGACGUUCACAUGCCCGGAUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGGCCAUCUUUUUUUUA____ .............................................................((((((((.((((........))))...((((((((.....))))))))((((.......)))))))))))).................... (-32.85 = -31.85 + -1.00)
| Location | 7,957,023 – 7,957,165 |
|---|---|
| Length | 142 |
| Sequences | 11 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 67.76 |
| Shannon entropy | 0.70249 |
| G+C content | 0.49468 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -29.18 |
| Energy contribution | -28.28 |
| Covariance contribution | -0.90 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
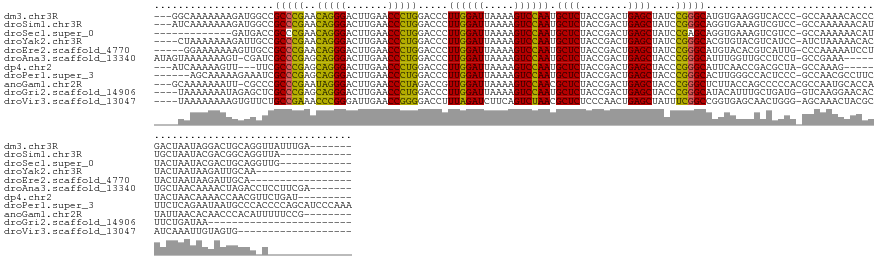
>dm3.chr3R 7957023 142 - 27905053 ---GGCAAAAAAAGAUGGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUGAAGGUCACCC-GCCAAAACACCCGACUAAUAGGACUGCAGGUUAUUUGA------- ---.........((((((((((((((.(((((.......))))).....))))......(((((...((((........))))....((((..(((...(((.....-)))...))))))).......))))))).))))))))..------- ( -42.20, z-score = -1.37, R) >droSim1.chr3R 14082390 137 + 27517382 ---AUCAAAAAAAGAUGGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAGGUGAAAGUCGUCC-GCCAAAAAACAUUGCUAAUACGACGGCAGGUUA------------ ---............((((.(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).((((.....)).))-))))...(((.(((((........)))))))).------------ ( -41.90, z-score = -1.65, R) >droSec1.super_0 7101578 127 + 21120651 -------------GAUGACCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGAGCAGGUGAAAGUCGUCC-GCCAAAAAACAUUACUAAUACGACUGCAGGUUG------------ -------------((((((((((....(((((.......)))))....(((((((............((((........)))).)))))))..))))...)))))).-....................((((.....))))------------ ( -31.92, z-score = -1.18, R) >droYak2.chr3R 7764400 132 - 28832112 ----CUAAAAAAAGAUUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCACGUGUACGUCAUCC-AUCUAAAAACACUACUAAUAAGAUUGCAA---------------- ----...........((((.(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))..((((.........-........)))).............))))---------------- ( -33.23, z-score = -2.42, R) >droEre2.scaffold_4770 14848456 130 + 17746568 -----GGAAAAAAAGUUGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUACACGUCAUUG-CCCAAAAAUCCUUACUAAUAAGAUUGCA----------------- -----(((............(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).((..((......))-..))....))).................----------------- ( -31.90, z-score = -1.25, R) >droAna3.scaffold_13340 5404152 139 - 23697760 AUAGUAAAAAAAGU-CGAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUUGGUUGCCUCCU-GCCGAAA-----UGCUAACAAAACUAGACCUCCUUCGA------- .(((((........-.....(((((..(((((.......))))).....((((((.....)))))).((((........))))....))))).((((((........-)))))).-----))))).....................------- ( -37.80, z-score = -1.70, R) >dp4.chr2 19721640 132 - 30794189 ---AUCAAAAAGUU---UUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUUCAACCGACGCUA-GCCAAAG-----UACUAACAAAACCAACGUUCUGAU--------- ---((((.((.(((---...(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))...........(((.-.....))-----)............))).)).))))--------- ( -30.90, z-score = -0.92, R) >droPer1.super_3 2464834 146 - 7375914 ------AGCAAAAAGAAAUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACUUGGGCCACUCCC-GCCAACGCCUUCUUCUCAGAAUAAUGCCCACCCCAGCAUCCCAAA ------.((...........(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))...(((((.......-((....)).((((....))))....))))).....))........ ( -40.10, z-score = -1.46, R) >anoGam1.chr2R 51278841 141 - 62725911 ---GCAAAAAAAUU-CGCCCGCCCGAAUAGGGACUUGAACCCUAGACCGUUGGAUUAAAAGUCCAACGCUCUACCGACUGAGCUACCCGGGCUCUUACCAGCCCCCACGCCAAUGCACCAUAUUAACACAACCCACAUUUUUCCG-------- ---(((........-.....(((((..(((((.......)))))....(((((((.....)))))))((((........))))....)))))........((......))...))).............................-------- ( -34.30, z-score = -3.67, R) >droGri2.scaffold_14906 11425882 124 - 14172833 ----UAAAAAAAUAGAGCUCGCCCGAGCAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUACAUUUGCUGAUG-GUCAAGGAACACUUCUGAUAA------------------------ ----........((((((((....)))).(...(((((.((((((....((((((.....)))))).((((........))))...))))(((......)))....)-))))))...)...))))....------------------------ ( -34.10, z-score = -1.71, R) >droVir3.scaffold_13047 1772746 129 - 19223366 ----UAAAAAAAAGUGUUCUGCCGAAACCCGGGAUUGAACCGGGGACCUUUAGAUCUUCAGUCUAACGCUCUCCCAACUGAGCUAUUUCGGCCGGUGAGCAACUGGG-AGCAAACUACGCAUCAAAUUGUAGUG------------------- ----..........((((((((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))((((.....))))))-)))).((((((........)))))).------------------- ( -42.30, z-score = -2.76, R) >consensus ____UAAAAAAAAGAUGGCCGCCCGAACAGGGACUUGAACCCUGGACCCUUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUAUCCGGGCAUGUGAACGUCGCCC_GCCAAAAAACACUACUAAUAAGACUGCAG_U______________ ....................(((((..(((((.......))))).....((((((.....)))))).((((........))))....)))))............................................................. (-29.18 = -28.28 + -0.90)
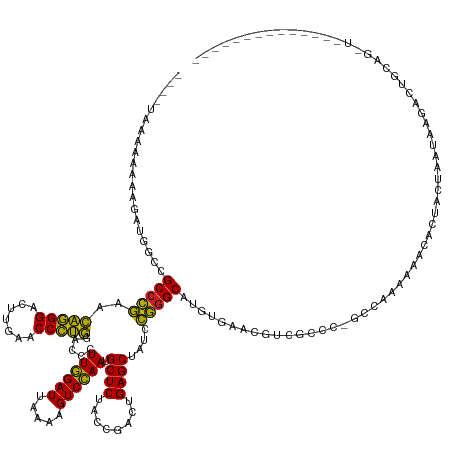
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:17 2011