| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,934,090 – 7,934,196 |
| Length | 106 |
| Max. P | 0.682217 |

| Location | 7,934,090 – 7,934,196 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.74 |
| Shannon entropy | 0.36744 |
| G+C content | 0.36861 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.16 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
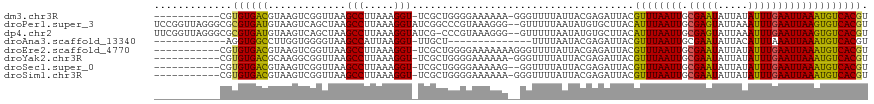
>dm3.chr3R 7934090 106 + 27905053 -----------CGUGUGACGUAAGUCGGUUAAGCCUUAAAGGU-UCGCUGGGGAAAAAA-GGGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU -----------...(((((((((.(((...(((((((.....(-((......)))...)-)))))).....)))..))))((((((((.((((((...))))))))))))))))))).. ( -26.60, z-score = -2.13, R) >droPer1.super_3 5479393 117 + 7375914 UCCGGUUAGGGCGCGUGAUGUAAGUCAGCUAAGCCUUAAAGGUAUCGGCCCGUAAAGGG--GUUUUUAAUAUGUGCUUACAUUUAAUUGCGAGUAUUAAAUUUGAAUUAAGUGUCACGU .((..(((((((((.((((....))))))...))))))).))((..(((((......))--)))..))..(((((...((((((((((.(((((.....)))))))))))))))))))) ( -32.60, z-score = -1.82, R) >dp4.chr2 22703274 116 + 30794189 UUCGGUUAGGGCGCGUGAUGUAAGUCAGCUAAGCCUUAAAGGUAUCG-CCCGUAAAGGG--GUUUUUAAUAUGUGCUUACAUUUAAUUGCGAGUAUUAAAUUUGAAUUAAGUGUCACGU .((..(((((((((.((((....))))))...))))))).))....(-(((......))--)).......(((((...((((((((((.(((((.....)))))))))))))))))))) ( -29.70, z-score = -1.31, R) >droAna3.scaffold_13340 19219810 92 - 23697760 ------------AGGUGGCCUUGGUGGGGUAAGCCAUUAAGGU-UUGCU--------------UUUUAAUACGAGAUUACGUUUAAUUGCGAAUAUUACAUUUAAAUUAAAUGUCACGU ------------..(((((((((((((......))))))))))-((((.--------------..((((.(((......)))))))..)))).....(((((((...)))))))))).. ( -23.10, z-score = -2.08, R) >droEre2.scaffold_4770 4057673 107 - 17746568 -----------CGUGUGACGUAAGUCGGUUAAGCCUUAAAGGU-UCGCUGGGGAAAAAAAGGGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU -----------...(((((((((.(((...(((((((.....(-((......)))....))))))).....)))..))))((((((((.((((((...))))))))))))))))))).. ( -27.00, z-score = -2.34, R) >droYak2.chr3R 12246327 106 + 28832112 -----------CGUGUGACGCAAGGCGGUUAAGCCUUAAAGGU-UCGCUGGGGAAAAAA-GGGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU -----------...((((((.(((((......)))))....((-((((....(((....-.((((((.....))))))...)))....)))))).................)))))).. ( -24.10, z-score = -1.00, R) >droSec1.super_0 7078797 105 - 21120651 -----------CGUGUGACGUAAGUCGGUUAAGCCUUAAAGGU-UCGCUGGGGAAAAAG--GGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU -----------...(((((((((.(((...(((((((.....(-((......)))..))--))))).....)))..))))((((((((.((((((...))))))))))))))))))).. ( -26.20, z-score = -1.98, R) >droSim1.chr3R 14059394 106 - 27517382 -----------CGUGUGACGUAAGUCGGUUAAGCCUUAAAGGU-UCGCUGGGGAAAAAA-GGGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU -----------...(((((((((.(((...(((((((.....(-((......)))...)-)))))).....)))..))))((((((((.((((((...))))))))))))))))))).. ( -26.60, z-score = -2.13, R) >consensus ___________CGUGUGACGUAAGUCGGUUAAGCCUUAAAGGU_UCGCUGGGGAAAAAA__GGUUUUAUUACGAGAUUACGUUUAAUUGCGAAUAUUAUAUUUGAAUUAAAUGUCACGU .............((((((.......(((...(((.....)))...)))...............................((((((((.(((((.....))))))))))))))))))). (-15.46 = -15.16 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:16 2011