| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,922,761 – 7,922,858 |
| Length | 97 |
| Max. P | 0.997162 |

| Location | 7,922,761 – 7,922,858 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.35 |
| Shannon entropy | 0.53801 |
| G+C content | 0.39110 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -8.94 |
| Energy contribution | -8.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
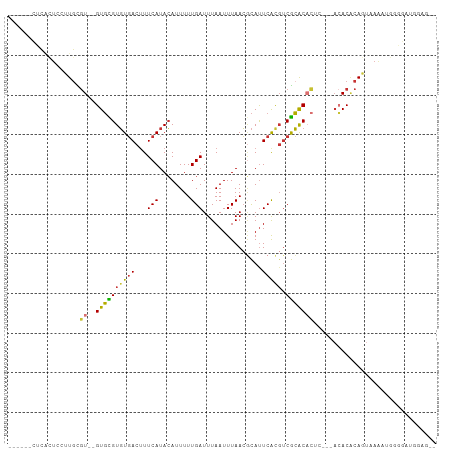
>dm3.chr3R 7922761 97 + 27905053 ------CUCACUCCUUGCGU--GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCACGUCGCACACUCUA-ACACACAGUUAACUGGGGAUGGAG-- ------....((((((.(((--(((((((((.....))))).................(((......))).))))))....-.....(((....)))).)).))))-- ( -24.60, z-score = -2.22, R) >droSim1.chr3R 14047482 91 - 27517382 ------CUCACGCCUUGCGU--GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCACGUCGCACAC-------ACACAGUAAAAUGGGGAAGGAG-- ------(((.(.((((((((--(((.(((((((..(((........)))...........(......)))))))).)-------)))).))))...)).)...)))-- ( -23.90, z-score = -1.62, R) >droSec1.super_0 7067283 97 - 21120651 ------CUCACUCCUUACGU--GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACACAUUCACGUCGCACACUCCG-ACACACAGUAAAAUGGGGAAGGAG-- ------....((((((..((--((((((((((...(((........)))...............))))).)))))))((((-((.....))....)))).))))))-- ( -23.27, z-score = -2.00, R) >droYak2.chr3R 12233496 96 + 28832112 ------CUCACUCCUUGCGU--GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCACGUCGCACA-UCUA-ACACGCAGUAAAAAGAAGAUGAAA-- ------.(((((.(((((((--((..(((((((..(((........)))...........(......)))))))).-....-)))))))......)).)).)))..-- ( -19.80, z-score = -0.73, R) >droEre2.scaffold_4770 4045660 97 - 17746568 ------CUCGCUCCUUGCGU--GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCACUUCGCACACUCUA-ACACACAGUUAAAAGAAGUUGGAA-- ------...((.....))((--(((((((((.....)))))....((((......))))............))))))((((-((...............)))))).-- ( -20.56, z-score = -1.20, R) >dp4.chr2 22690136 87 + 30794189 AUCUUUCUUUUUGUGUGUGU--GUGUGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCGUGUCGCACUC-----ACACACACUCAA-------------- ..........(((.((((((--(((.(((((((..(((........))).........(((....)))))))))).)-----)))))))).)))-------------- ( -30.20, z-score = -5.54, R) >droPer1.super_3 5466226 83 + 7375914 ----AUCUUUCUUUUUGUGU--GUGUGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCAUGUCGCACUC-----ACACACACUCAA-------------- ----...........(((((--(((.(((((((..(((........)))...................))))))).)-----))))))).....-------------- ( -22.70, z-score = -4.31, R) >droGri2.scaffold_14830 1763576 102 + 6267026 ------UUCAGUGUGUGUAUUGGUCUGUUUCACAUUCAUAAAUUUUUGAUUUAAUUUAAGUUACUCAUGCCAGGCAUUCAGCAGACUCACACAGACCUGACCUAAGAU ------.(((((.(((((...((((((((.........((((((........))))))........((((...))))..)))))))).))))).).))))........ ( -21.10, z-score = -0.79, R) >consensus ______CUCACUCCUUGCGU__GUGCGUGUGACUUUCAUACAUUUUUGAUUUAAUUUAACGCAUUCACGUCGCACACUC___ACACACAGUAAAAUGGGGAUGGAG__ ......................((((((((((...(((........)))...............))))).)))))................................. ( -8.94 = -8.38 + -0.56)


| Location | 7,922,761 – 7,922,858 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.35 |
| Shannon entropy | 0.53801 |
| G+C content | 0.39110 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
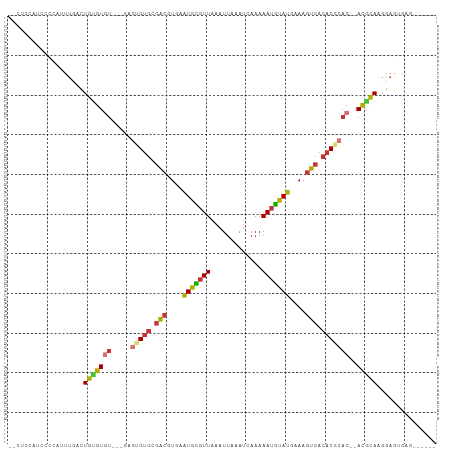
>dm3.chr3R 7922761 97 - 27905053 --CUCCAUCCCCAGUUAACUGUGUGU-UAGAGUGUGCGACGUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC--ACGCAAGGAGUGAG------ --((((......((....))((((((-....(((((.(((....(((((((.............)))))))....))).)))))))--))))..))))....------ ( -27.02, z-score = -2.64, R) >droSim1.chr3R 14047482 91 + 27517382 --CUCCUUCCCCAUUUUACUGUGU-------GUGUGCGACGUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC--ACGCAAGGCGUGAG------ --.............((((.((((-------(((((.(((....(((((((.............)))))))....))).)))))))--))((...)))))).------ ( -26.32, z-score = -2.56, R) >droSec1.super_0 7067283 97 + 21120651 --CUCCUUCCCCAUUUUACUGUGUGU-CGGAGUGUGCGACGUGAAUGUGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC--ACGUAAGGAGUGAG------ --((((((((.(((........))).-.)))(((((((..((((.........(((........))).........)))).)))))--))...)))))....------ ( -24.17, z-score = -1.53, R) >droYak2.chr3R 12233496 96 - 28832112 --UUUCAUCUUCUUUUUACUGCGUGU-UAGA-UGUGCGACGUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC--ACGCAAGGAGUGAG------ --..(((.(((((......(((((((-..(.-((((.(((....(((((((.............)))))))....))).)))))))--))))))))))))).------ ( -26.72, z-score = -1.97, R) >droEre2.scaffold_4770 4045660 97 + 17746568 --UUCCAACUUCUUUUAACUGUGUGU-UAGAGUGUGCGAAGUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC--ACGCAAGGAGCGAG------ --((((.......((((((.....))-))))(((((((..(((((((((((.............))))))).....)))).)))))--))....))))....------ ( -24.52, z-score = -1.51, R) >dp4.chr2 22690136 87 - 30794189 --------------UUGAGUGUGUGU-----GAGUGCGACACGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACACAC--ACACACACAAAAAGAAAGAU --------------(((.((((((((-----(.(((.(((....(((((((.............)))))))....))).))).)))--)))))).))).......... ( -29.12, z-score = -5.09, R) >droPer1.super_3 5466226 83 - 7375914 --------------UUGAGUGUGUGU-----GAGUGCGACAUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACACAC--ACACAAAAAGAAAGAU---- --------------.....(((((((-----(.(((.(((.(..(((((((.............)))))))..).))).))).)))--)))))...........---- ( -24.62, z-score = -4.15, R) >droGri2.scaffold_14830 1763576 102 - 6267026 AUCUUAGGUCAGGUCUGUGUGAGUCUGCUGAAUGCCUGGCAUGAGUAACUUAAAUUAAAUCAAAAAUUUAUGAAUGUGAAACAGACCAAUACACACACUGAA------ ...........(((((((.(((((.((((..((((...)))).)))))))))..............(((((....))))))))))))...............------ ( -20.80, z-score = -0.42, R) >consensus __CUCCAUCCCCAUUUGACUGUGUGU___GAGUGUGCGACGUGAAUGCGUUAAAUUAAAUCAAAAAUGUAUGAAAGUCACACGCAC__ACGCAAGGAGUGAG______ ..................((.(((((.....(((((.(((....(((((((.............)))))))....))).)))))....))))).))............ (-14.11 = -14.38 + 0.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:15 2011