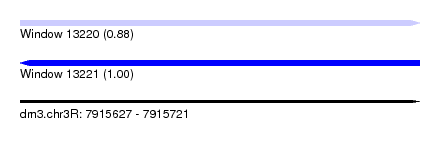
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,915,627 – 7,915,721 |
| Length | 94 |
| Max. P | 0.995584 |

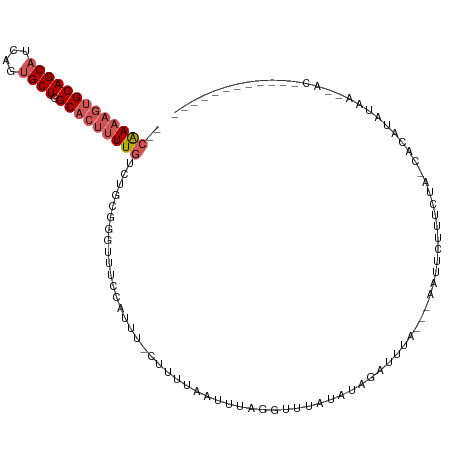
| Location | 7,915,627 – 7,915,721 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 62.89 |
| Shannon entropy | 0.63802 |
| G+C content | 0.34501 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -11.86 |
| Energy contribution | -12.90 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
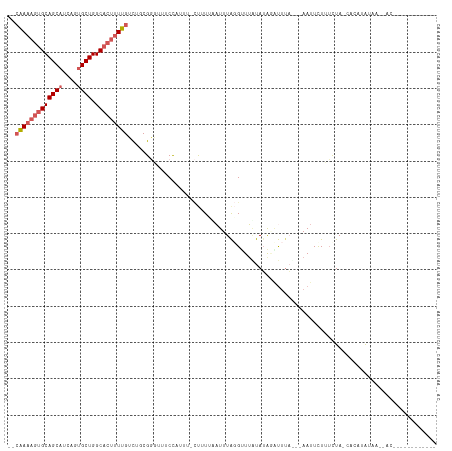
>dm3.chr3R 7915627 94 + 27905053 --CAAAAGUGCAGCCUCAGUGCUGGCACUUUUGUUUUCGG-UUUCCAUCA-CUAUUAUUUUAGGCUAAUAUAAAUUCAUUGAAUUCUUUCUA-UAGUUA------------------- --((((((((((((......))).))))))))).......-.........-...........(((((..(..(((((...)))))..)....-))))).------------------- ( -16.00, z-score = -0.89, R) >droEre2.scaffold_4770 4038359 107 - 17746568 CAAGACAAGGCAGCAUCAGUGCUGGCACUUUUGGCUUCGCUUUUCUUUUC--CCUAAAUUUACGCUUUUAAGUGUCUUA--AAUUCUGUUUA-UAAAAAUACUUACUCAUUA------ .(((((..(.(((((....))))).)((((..(((...............--...........)))...))))))))).--...........-...................------ ( -16.67, z-score = -0.14, R) >droYak2.chr3R 12226100 114 + 28832112 --CAAAAGUGCAGCAUCAGUGCUGGCACCUUUGGCUCCGCUUUUCUUUUUAUUUUUCAUUUGCGUUUGUAUAUGCUUUC--AAUUCUGGCUAUGGUCAACAAAGACACCGAUUUAUUA --((((.((((((((....)))).)))).))))....((....(((((.............((((......))))....--.....((((....))))..)))))...))........ ( -20.20, z-score = 0.69, R) >droSec1.super_0 7060100 106 - 21120651 --CAAAAGUGCAGCAUCAGUGCUGGCACUUUUGUUUGCGGGUUUCCAUUU-CUUUUCAUUUAGGUUUAUAUAGCUUUA---AAUUUUUUCUAACACAUAUACUGACAUGUUA------ --(((((((((((((....)))).))))))))).....((....))....-......(((((((((.....))).)))---)))............................------ ( -18.30, z-score = -0.38, R) >droSim1.chr3R 14040269 96 - 27517382 --CAAAAGUGCAGCAUCAGUGCUGGCACUUUUGUCUGCGGGUUUCCAUUU-CUUUCUAUUUAGGUUUAUAUAGAUUUA---AAUUCUUUUUAACACAUGUUA---------------- --(((((((((((((....)))).))))))))).....((....))....-......((((((((((....)))))))---)))......((((....))))---------------- ( -20.60, z-score = -2.09, R) >consensus __CAAAAGUGCAGCAUCAGUGCUGGCACUUUUGUCUGCGGGUUUCCAUUU_CUUUUAAUUUAGGUUUAUAUAGAUUUA___AAUUCUUUCUA_CACAUAUAA__AC____________ ..(((((((((((((....)))).)))))))))..................................................................................... (-11.86 = -12.90 + 1.04)

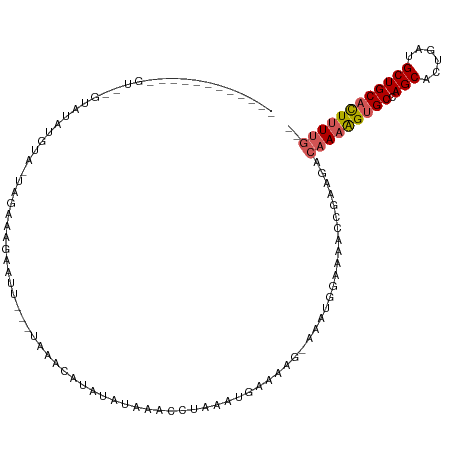
| Location | 7,915,627 – 7,915,721 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 62.89 |
| Shannon entropy | 0.63802 |
| G+C content | 0.34501 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7915627 94 - 27905053 -------------------UAACUA-UAGAAAGAAUUCAAUGAAUUUAUAUUAGCCUAAAAUAAUAG-UGAUGGAAA-CCGAAAACAAAAGUGCCAGCACUGAGGCUGCACUUUUG-- -------------------...(((-(.....((((((...)))))).(((((........))))).-..))))...-.......(((((((((.(((......))))))))))))-- ( -18.80, z-score = -2.53, R) >droEre2.scaffold_4770 4038359 107 + 17746568 ------UAAUGAGUAAGUAUUUUUA-UAAACAGAAUU--UAAGACACUUAAAAGCGUAAAUUUAGG--GAAAAGAAAAGCGAAGCCAAAAGUGCCAGCACUGAUGCUGCCUUGUCUUG ------......(((((....))))-)..........--((((((.((((((........))))))--........(((.(.(((....((((....))))...))).)))))))))) ( -16.20, z-score = 0.78, R) >droYak2.chr3R 12226100 114 - 28832112 UAAUAAAUCGGUGUCUUUGUUGACCAUAGCCAGAAUU--GAAAGCAUAUACAAACGCAAAUGAAAAAUAAAAAGAAAAGCGGAGCCAAAGGUGCCAGCACUGAUGCUGCACUUUUG-- .......((((((((......)))....)))......--...............(((.....................)))))..(((((((((.((((....)))))))))))))-- ( -21.80, z-score = -0.13, R) >droSec1.super_0 7060100 106 + 21120651 ------UAACAUGUCAGUAUAUGUGUUAGAAAAAAUU---UAAAGCUAUAUAAACCUAAAUGAAAAG-AAAUGGAAACCCGCAAACAAAAGUGCCAGCACUGAUGCUGCACUUUUG-- ------...(((.....(((((((.(((((.....))---))).)).))))).......))).....-....((....)).....(((((((((.((((....)))))))))))))-- ( -21.00, z-score = -2.38, R) >droSim1.chr3R 14040269 96 + 27517382 ----------------UAACAUGUGUUAAAAAGAAUU---UAAAUCUAUAUAAACCUAAAUAGAAAG-AAAUGGAAACCCGCAGACAAAAGUGCCAGCACUGAUGCUGCACUUUUG-- ----------------.....((((.........(((---(...(((((.((....)).)))))...-))))(....).))))..(((((((((.((((....)))))))))))))-- ( -21.50, z-score = -4.00, R) >consensus ____________GU__GUAUAUGUA_UAGAAAGAAUU___UAAACAUAUAUAAACCUAAAUGAAAAG_AAAUGGAAAACCGAAGACAAAAGUGCCAGCACUGAUGCUGCACUUUUG__ .....................................................................................(((((((((.(((......)))))))))))).. (-12.32 = -12.80 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:13 2011