
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,908,561 – 7,908,658 |
| Length | 97 |
| Max. P | 0.603362 |

| Location | 7,908,561 – 7,908,658 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.07 |
| Shannon entropy | 0.67032 |
| G+C content | 0.50783 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7908561 97 - 27905053 GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCCAUUCCC--CCUCUUCCA-------GGCAGCCU-UCGGCGGCUCGACAAGGCCGCCAUAUGGAGGAGGU ...((((.....(((((...............)))))..))))..--(((((((((-------((...)).-..(((((((......)))))))...))))))))). ( -30.66, z-score = -1.67, R) >droSim1.chr3R 14033229 97 + 27517382 GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCCAUUCCC--ACUCUUCCA-------GGCAGCCU-UCGGCGGCUCGACAAGGCCGCCAUAUGGAGGAGGU ...((((.....(((((...............)))))..))))..--.((((((((-------((...)).-..(((((((......)))))))...)))))))).. ( -28.26, z-score = -1.18, R) >droSec1.super_0 7053097 97 + 21120651 GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCCAUUCCC--ACUCUUCCA-------GGCAGCCU-UCGGCGGCUCGACAAGGCCGCCAUAUGGAGGAGGU ...((((.....(((((...............)))))..))))..--.((((((((-------((...)).-..(((((((......)))))))...)))))))).. ( -28.26, z-score = -1.18, R) >droYak2.chr3R 12218885 96 - 28832112 GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCCAUUCCC---CUCUUCCA-------GGCAGCCU-UCGGCGGCUCGACAAGGCCGCCAUAUGGAGGAGGU ...((((.....(((((...............)))))..)))).(---((((((((-------((...)).-..(((((((......)))))))...))))))))). ( -30.66, z-score = -1.70, R) >droEre2.scaffold_4770 4031420 97 + 17746568 GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCUAUUACC--CCUCUUCCA-------GGCAGCCU-UCGGCGGCUCGACAAGGCCGCCAUAUGGAGGAGGU ............(((((...............)))))........--(((((((((-------((...)).-..(((((((......)))))))...))))))))). ( -30.36, z-score = -1.99, R) >droAna3.scaffold_13340 19194194 92 + 23697760 GACGAAUCUUUUGAAUGCUUACUGACUUCAGCUAUCCUCUCUC-----GUCCCCCA---------CAGCCA-UCGGCGGCGCGCCAAGGCCGACACAUGGCGGCGGU (((((.((....)).......(((....)))..........))-----)))..((.---------(.((((-(.(.((((.(.....)))))...)))))).).)). ( -24.10, z-score = 0.74, R) >dp4.chr2 22673053 103 - 30794189 GACGAAUCAUUUGAAUGCUUACUGAAUGCUUCCACUCUCCUCCUUUGACUCUUCCCCUGUUCCUUUAGCCU-ACGGCGGCUCGACAAGGCCAACACAUGGAGGU--- ...(((.(((((...........))))).)))......(((((..((....................(((.-..)))((((......))))....)).))))).--- ( -20.20, z-score = 0.31, R) >droPer1.super_3 5448041 103 - 7375914 GACGAAUCAUUUGAAUGCUUACUGAAUGCUUCCACUCUCCUCCUUUGACUCUUCCUCUGUUCCUUUAGCCU-ACGGCGGCUCGACAAGGCCAACACAUGGAGGU--- ...(((.(((((...........))))).)))......(((((..((....................(((.-..)))((((......))))....)).))))).--- ( -20.20, z-score = 0.25, R) >droWil1.scaffold_181096 1694621 88 - 12416693 ----AAUGAUUCAUAUUUUGA--AUGCUUAG---UGAAUCUCUCUCUCACCUUU---------UGCAGUUGAACGGCUACA-CACAAAGCCAACACAUGGAGGUUUA ----....(((((.....)))--))......---(((((((((...(((.((..---------...)).)))..((((...-.....)))).......))))))))) ( -16.30, z-score = -0.33, R) >droGri2.scaffold_14853 1840831 98 + 10151454 CAUGAUUAAUCUGUAUACUAA--AAAUUCAAUUUUGGAUUUUCCUGUUGUCUCUGUG---UUGUAUAGCUG-ACGGCGGCAGGGCAAGGCCAACACAUGGAGUU--- ((((................(--(((((((....))))))))..(((((.(..(((.---((((...((..-...)).)))).)))..).))))))))).....--- ( -23.30, z-score = -0.42, R) >consensus GACGAAUCAUUUGAAUGCUUACUGAAUCCACACAUUCCCAUUCCC__ACUCUUCCA_______GGCAGCCU_UCGGCGGCUCGACAAGGCCGACACAUGGAGGAGGU ..................................................(((((............(((....)))(((........))).......))))).... (-11.81 = -12.01 + 0.20)
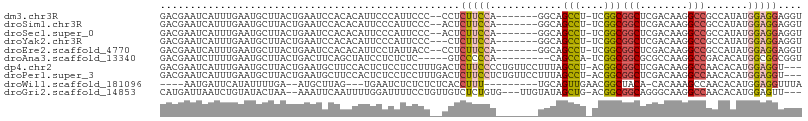
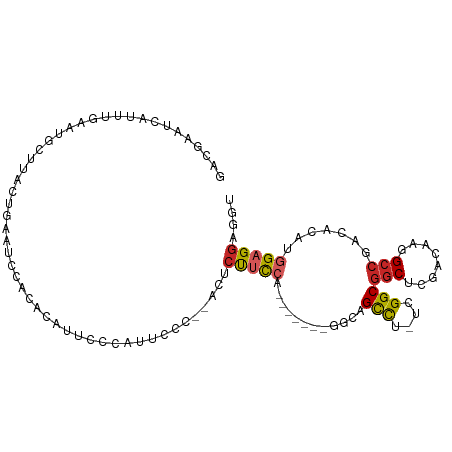

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:12 2011