
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,878,038 – 7,878,145 |
| Length | 107 |
| Max. P | 0.514669 |

| Location | 7,878,038 – 7,878,145 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.34391 |
| G+C content | 0.43752 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -13.86 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
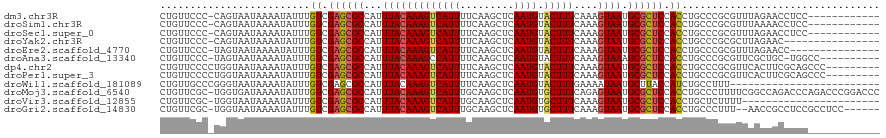
>dm3.chr3R 7878038 107 + 27905053 CUGUUCCC-CAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUUAGAACCUCC------------ ..((((..-................((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))..(....).....))))....------------ ( -16.90, z-score = -1.49, R) >droSim1.chr3R 14001522 107 - 27517382 CUGUUCCC-CAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUUAAAACCUCC------------ (((.....-))).............((.((((((...(((((((((((((.........)))).)))))....)))).)))))).)).....................------------ ( -15.90, z-score = -1.60, R) >droSec1.super_0 7022962 107 - 21120651 CUGUUCCC-CAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUUAGAACCUCC------------ ..((((..-................((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))..(....).....))))....------------ ( -16.90, z-score = -1.49, R) >droYak2.chr3R 12188194 103 + 28832112 CUGUUCCC-CAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGCUUAGAAC---------------- ..((((..-................((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))..((....))...))))---------------- ( -17.50, z-score = -1.24, R) >droEre2.scaffold_4770 4001091 104 - 17746568 CUGUUCCC-UAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUUAGAACC--------------- ..((((..-((....))........((.((((((...(((((((((((((.........)))).)))))....)))).)))))).)).............)))).--------------- ( -17.10, z-score = -1.32, R) >droAna3.scaffold_13340 19166034 108 - 23697760 CUGUUCCC-UAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGCCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAAGCGCUCCACCUGCCCGCGUUCGCUGC-UGGCC---------- ........-................((.((((((..(((((..(((.........)))...((......))..))))))))))).))..(((.(((.....)))-.))).---------- ( -20.30, z-score = -0.37, R) >dp4.chr2 22637259 111 + 30794189 CUGUUCCCCUGGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUCACUUCGCAGCCC--------- ..........(((............((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))......(((.......))).))).--------- ( -21.60, z-score = -1.82, R) >droPer1.super_3 5412503 111 + 7375914 CUGUUCCCCUGGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUCACUUCGCAGCCC--------- ..........(((............((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))......(((.......))).))).--------- ( -21.60, z-score = -1.82, R) >droWil1.scaffold_181089 475248 95 + 12369635 CUGUUGCCCGGGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUGAAAAUAAUGCUUACCAUCUGCCUUU------------------------- .((((((....)))))).......((..(((((..((((.((((((((((.........)))).)))))).))))..)))))..)).........------------------------- ( -18.10, z-score = -1.26, R) >droMoj3.scaffold_6540 24363257 119 + 34148556 CUGUUCGC-UGGUGAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUGCAAGCUCAAUGUGCUUUCAGAGUAAUGCGCUCCACCUGCCCUUUUCGGCCAGACCCAGACCCGGACCC ..((((((-(((.(((((.....)))))((((((...((((.......((((.((((.......)))).)))))))).))))))...(((.((.....)).)))..))))...))))).. ( -31.61, z-score = -1.49, R) >droVir3.scaffold_12855 9271248 96 + 10161210 CUGUUCGC-UGGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUGCAAGCUCAAUGUGCUUUCAAAGUAAUGCGCUCCACCUGCUCUUUU----------------------- ......((-.(((.((((.....)))).((((((...((((.......((((.((((.......)))).)))))))).)))))).))).))......----------------------- ( -20.51, z-score = -0.97, R) >droGri2.scaffold_14830 1741241 111 + 6267026 CUGUUCGC-UGGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUGCAAGCUCAAUGUGCUUUCAAAGUAAUGCGCUCCACCUGCCCUUU--AACCGCCUCCGCCUCC------ ......((-.(((..((((......((.((((((...((((.......((((.((((.......)))).)))))))).)))))).))......)))--))))))..........------ ( -21.21, z-score = -0.90, R) >consensus CUGUUCCC_UAGUAAUAAAAUAUUUGUCGAGCGCCAUUUACAAAGUCAUUUUCAAGCUCAAUGUACUUUCAAAGUAAUGCGCUCCACCUGCCCGCGUUUACAACCUCC____________ .........................((.((((((...(((((((((((((.........)))).)))))....)))).)))))).))................................. (-13.86 = -13.88 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:07 2011