| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,875,509 – 7,875,602 |
| Length | 93 |
| Max. P | 0.542473 |

| Location | 7,875,509 – 7,875,602 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 68.27 |
| Shannon entropy | 0.65503 |
| G+C content | 0.41871 |
| Mean single sequence MFE | -20.09 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
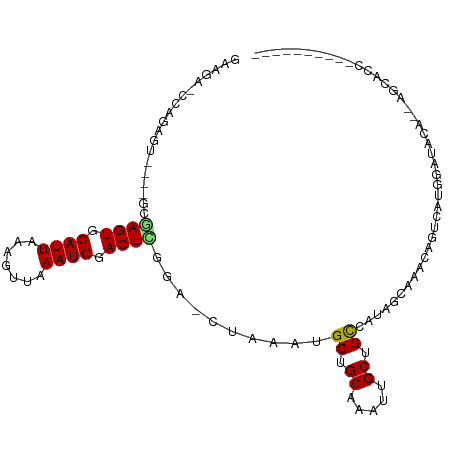
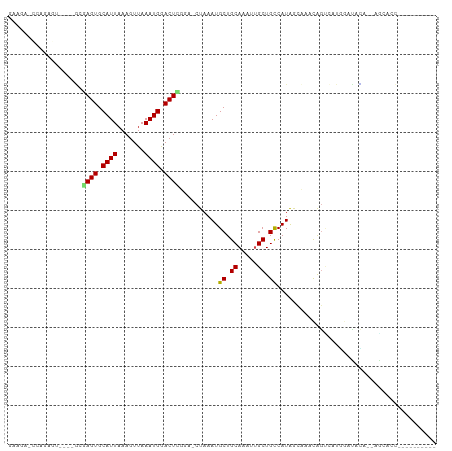
>dm3.chr3R 7875509 93 + 27905053 GUAGAGCCAGAGU----GCGAGUGCAUUAAAGUUAAAUGGACUCGGA-CUAAAUGCUGCAAAUUGCUGCCAUAGCCAACAGUCAUGGAUACA--AACACC---------- ((((......(((----.(((((.((((.......)))).))))).)-)).....))))...(((...((((..(.....)..))))...))--).....---------- ( -25.40, z-score = -1.76, R) >droSim1.chr3R 13998966 93 - 27517382 GAAGAGCCAGACU----GCGAGUGCAUUAAAGUUAAAUGGACUCGGA-CUAAAUGCUGCAAAUUGCUGCCAUAGCCAACAGUCAUGGAUACA--AGCACC---------- ......(((((((----(.((((.((((.......)))).))))((.-(((...((.((.....)).))..)))))..))))).))).....--......---------- ( -26.40, z-score = -2.07, R) >droSec1.super_0 7020423 93 - 21120651 GAAGAGCCAGAGU----GCGAGUGCAUUAAAGUUAAAUGGACUCGGA-CUAAAUGCUGCAAAUUGCUGCCAUAGCCAACAGUCAUGGAUACA--AGCACC---------- ....(((...(((----.(((((.((((.......)))).))))).)-))....)))......((((.((((..(.....)..)))).....--))))..---------- ( -26.10, z-score = -1.83, R) >droYak2.chr3R 12185713 93 + 28832112 GAAACCCCAGAGU----GAGAGUGCAUUAAAGUUAAAUGGACUCGUA-CUAAAUGCUGCAAAUUGCUGCCAUAGCCAACAGUCAUGGAUACA--AGCACC---------- ..........(((----(.((((.((((.......)))).)))).))-))...((((((.....))..((((..(.....)..)))).....--))))..---------- ( -20.00, z-score = -0.79, R) >droEre2.scaffold_4770 3998652 93 - 17746568 GAAGACCCAGAGU----GAGAGUGCAUUAAAGUUAAAUGGACUCGUA-CUAAAUGCUGCAAAUUGCUGCCAUAGCCAGCAGUCAUGGAUACA--AGCACC---------- ..........(((----(.((((.((((.......)))).)))).))-))...((((....((((((((....).)))))))..((....))--))))..---------- ( -21.10, z-score = -0.61, R) >droAna3.scaffold_13340 19163079 93 - 23697760 GCAGACCAAGAGCUGAAGUCAGUGCAUUAAAGUUAAAUGGACUCGGGUCUAAAUGCUGCAAAUUGCUGCCAUAACCAAUGUCCAACAGUCUAC----------------- ..((((...(.((((....)))).)............(((((...(((......((.((.....)).))....)))...)))))...))))..----------------- ( -21.90, z-score = -0.68, R) >dp4.chr2 22634822 94 + 30794189 GAAAAUAGAGAUG----GAGAGUGCAUUAAAGUUAAAUGGACUUGGA-CUAAAUGCUGCAAAUAGCUGCCAUAACAAACAGU-AUGGACACACCAGAAUC---------- .......((..((----(.((((.((((.......)))).))))...-......((((....))))(((((((........)-)))).))..)))...))---------- ( -15.40, z-score = -0.38, R) >droPer1.super_3 5410066 94 + 7375914 GAAAAUAGAGAUG----GAGAGUGCAUUAAAGUUAAAUGGACUUGGA-CUAAAUGCUGCAAAUAGCUGCCAUAACAAACAGU-AUGGACACACCAGAAAC---------- ...........((----(.((((.((((.......)))).))))...-......((((....))))(((((((........)-)))).))..))).....---------- ( -14.90, z-score = -0.56, R) >droWil1.scaffold_181089 11891455 80 - 12369635 ----------------GACAAGUGCAUUAAAGUUAAAUGGACUUG-GGCUAAAUGCUGCAAAUUGCUGCCAUAACAAACAGAAACAAAUACAAAUAC------------- ----------------..(((((.((((.......)))).)))))-(((.....((........)).)))...........................------------- ( -14.40, z-score = -1.15, R) >droVir3.scaffold_12855 9267847 91 + 10161210 -----------------UGCAGUGCAUUAAAGUUAAAUGGACUUUGCACUAAAUGCUGCAAAUUGCUGCCAUAGCAAACAGCAUACGACGGCCAUGUGUAUACUCGUU-- -----------------.((((((((...(((((.....)))))))))))..((((((....((((((...)))))).))))))......))................-- ( -21.60, z-score = 0.35, R) >droMoj3.scaffold_6540 24360412 89 + 34148556 -----------------UGCAGUGCAUUACACUCAAAUGGACUUUGCACUAAAUGCUGCAAAUUGCUGCCAUGAUAAACAG-AGAU-ACGACUGUGUGUACUCUCGUU-- -----------------.(((((.((((.......))))...(((((((.....).))))))..))))).......(((((-((((-(((....))))).)))).)))-- ( -18.40, z-score = 0.22, R) >droGri2.scaffold_14830 1738979 93 + 6267026 -----------------ACGAGUGCAUUAAAGUUAAAUGGACUUUCGACUAAAUGCUGCAAAUUGCUGUCAUAGCAAACAAAAACACGUAUACAUUUCCAGUUGCAGAGA -----------------..((((.((((.......)))).))))...........((((((.((((((...))))))((........))............))))))... ( -15.50, z-score = 0.57, R) >consensus GAAGA_CCAGAGU____GCGAGUGCAUUAAAGUUAAAUGGACUCGGA_CUAAAUGCUGCAAAUUGCUGCCAUAGCAAACAGUCAUGGAUACA__AGCACC__________ ...................((((.((((.......)))).))))..........((.((.....)).))......................................... ( -9.42 = -9.40 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:06 2011