| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,853,305 – 7,853,405 |
| Length | 100 |
| Max. P | 0.994124 |

| Location | 7,853,305 – 7,853,405 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.52163 |
| G+C content | 0.41021 |
| Mean single sequence MFE | -16.85 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.01 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
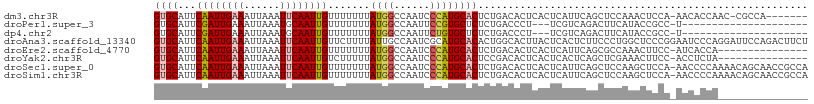
>dm3.chr3R 7853305 100 + 27905053 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUUUUUUAUGGCCAAUCCCAUGCACUCUGACACUCACUCAUUCAGCUCCAAACUCCA-AACACCAAC-CGCCA------- ((((...((((((((......)))))))).......((((......)))))))).((((...........))))............-.........-.....------- ( -14.90, z-score = -2.20, R) >droPer1.super_3 5385711 84 + 7375914 GUGCAUUCGAUUGAAAUUAAAUGCAAUUGUUUUUUUAUGGCCAAUUCCGUGCUCUCUGACCCU---UCGUCAGACUUCAUACCGCC-U--------------------- .((((((..((....))..)))))).............(((.......(((...((((((...---..))))))...)))...)))-.--------------------- ( -16.40, z-score = -1.83, R) >dp4.chr2 22610480 84 + 30794189 GUGCAUUCGAUUGAAAUUAAAUGCAAUUGUUUUUUUAUGGCCAAUUCUGUGCUCUCUGACCCU---UCGUCAGACUUCAUACCGCC-U--------------------- .((((((..((....))..)))))).............(((......((((...((((((...---..))))))...))))..)))-.--------------------- ( -17.60, z-score = -2.29, R) >droAna3.scaffold_13340 19140250 109 - 23697760 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUCUUUUAUUGCCAAUCGCAUGCACACUGGCACUUACUCACUCUUCCCUGGCUCCCGGAAUCCCAGGAUUCCAGACUUCU (((((..((((((((......)))))))).........(((.....))))))))......................((((.(((.((....)).)))..))))...... ( -24.80, z-score = -2.61, R) >droEre2.scaffold_4770 3975773 93 - 17746568 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUUUUUUAUGGCCAAUCCCAUGCACUCUGACACUCACUCAUUCAGCGCCAAACUUCC-AUCACCA--------------- ((((...((((((((......)))))))).......((((......)))))))).((((...........))))............-.......--------------- ( -14.90, z-score = -2.08, R) >droYak2.chr3R 12163329 93 + 28832112 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUCUUUUUAUGGCCAAUCCCAUGCACUCCGACACUCACUCACUCAGCUCGAAACUUCC-ACCUCUA--------------- ((((...((((((((......)))))))).......((((......))))))))................................-.......--------------- ( -14.20, z-score = -2.65, R) >droSec1.super_0 6998612 108 - 21120651 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUUUUUUAUGGCCAAUCCCAUGCACUCUGACACUCACUCAUUCAGCUCCAAGCUCCA-AACCCCAAAACAGCAACCGCCA ((((...((((((((......)))))))).......((((......))))))))..................(((.....)))...-............((....)).. ( -16.00, z-score = -1.49, R) >droSim1.chr3R 13976911 108 - 27517382 GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUUUUUUAUGGCCAAUCCCAUGCACUCUGACACUCACUCAUUCAGCUCCAAGCUCCA-AACCCCAAAACAGCAACCGCCA ((((...((((((((......)))))))).......((((......))))))))..................(((.....)))...-............((....)).. ( -16.00, z-score = -1.49, R) >consensus GUGCAUUCAAUUGAAAUUAAAUUCAAUUGUUUUUUUAUGGCCAAUCCCAUGCACUCUGACACUCACUCAUUCAGCUCCAAACUCCC_AACCCCA_______________ ((((...((((((((......)))))))).......((((......))))))))....................................................... (-10.73 = -11.01 + 0.28)
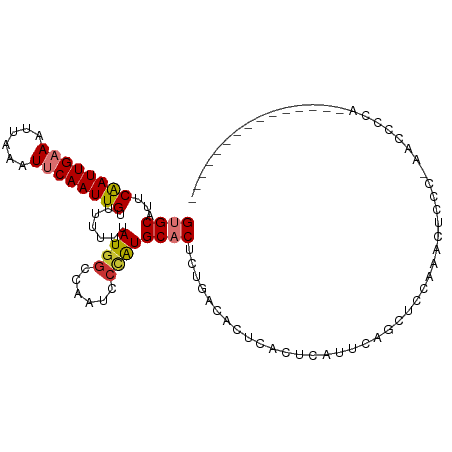
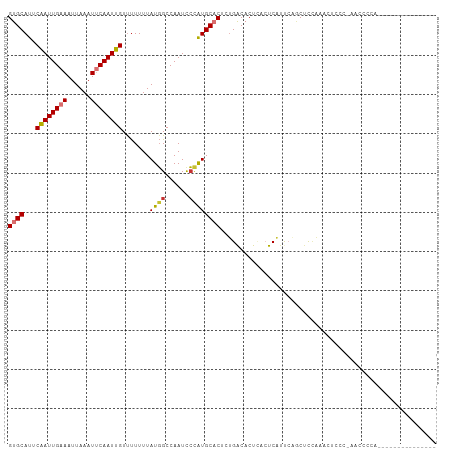
| Location | 7,853,305 – 7,853,405 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.52163 |
| G+C content | 0.41021 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -9.59 |
| Energy contribution | -10.96 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 7853305 100 - 27905053 -------UGGCG-GUUGGUGUU-UGGAGUUUGGAGCUGAAUGAGUGAGUGUCAGAGUGCAUGGGAUUGGCCAUAAAAAAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC -------..(((-((..((.((-((.(.((((..(((.(.....).)))..)))).).)).)).))..))).........((((((((......))))))))...)).. ( -26.80, z-score = -2.54, R) >droPer1.super_3 5385711 84 - 7375914 ---------------------A-GGCGGUAUGAAGUCUGACGA---AGGGUCAGAGAGCACGGAAUUGGCCAUAAAAAAACAAUUGCAUUUAAUUUCAAUCGAAUGCAC ---------------------.-(((.(..((...((((((..---...))))))...))......).))).............((((((..((....))..)))))). ( -18.40, z-score = -1.38, R) >dp4.chr2 22610480 84 - 30794189 ---------------------A-GGCGGUAUGAAGUCUGACGA---AGGGUCAGAGAGCACAGAAUUGGCCAUAAAAAAACAAUUGCAUUUAAUUUCAAUCGAAUGCAC ---------------------.-(((.(..((...((((((..---...)))))).....))....).))).............((((((..((....))..)))))). ( -18.40, z-score = -1.61, R) >droAna3.scaffold_13340 19140250 109 + 23697760 AGAAGUCUGGAAUCCUGGGAUUCCGGGAGCCAGGGAAGAGUGAGUAAGUGCCAGUGUGCAUGCGAUUGGCAAUAAAAGAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC .....(((((..((((((....)))))).))))).........(((..(((((((((....)).))))))).........((((((((......))))))))..))).. ( -33.50, z-score = -3.07, R) >droEre2.scaffold_4770 3975773 93 + 17746568 ---------------UGGUGAU-GGAAGUUUGGCGCUGAAUGAGUGAGUGUCAGAGUGCAUGGGAUUGGCCAUAAAAAAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC ---------------.......-.....(((((((((.(.....).)))))))))((((((((......)).........((((((((......)))))))).)))))) ( -25.30, z-score = -2.75, R) >droYak2.chr3R 12163329 93 - 28832112 ---------------UAGAGGU-GGAAGUUUCGAGCUGAGUGAGUGAGUGUCGGAGUGCAUGGGAUUGGCCAUAAAAAGACAAUUGAAUUUAAUUUCAAUUGAAUGCAC ---------------.......-......((((((((.(.....).))).)))))((((((((......)).........((((((((......)))))))).)))))) ( -20.20, z-score = -1.20, R) >droSec1.super_0 6998612 108 + 21120651 UGGCGGUUGCUGUUUUGGGGUU-UGGAGCUUGGAGCUGAAUGAGUGAGUGUCAGAGUGCAUGGGAUUGGCCAUAAAAAAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC (((((.(..((((((..((.(.-...).))..))))......))..).)))))..((((((((......)).........((((((((......)))))))).)))))) ( -30.30, z-score = -2.30, R) >droSim1.chr3R 13976911 108 + 27517382 UGGCGGUUGCUGUUUUGGGGUU-UGGAGCUUGGAGCUGAAUGAGUGAGUGUCAGAGUGCAUGGGAUUGGCCAUAAAAAAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC (((((.(..((((((..((.(.-...).))..))))......))..).)))))..((((((((......)).........((((((((......)))))))).)))))) ( -30.30, z-score = -2.30, R) >consensus _______________UGGGGUU_GGGAGUUUGGAGCUGAAUGAGUGAGUGUCAGAGUGCAUGGGAUUGGCCAUAAAAAAACAAUUGAAUUUAAUUUCAAUUGAAUGCAC .......................................................((((((((......)).........((((((((......)))))))).)))))) ( -9.59 = -10.96 + 1.38)

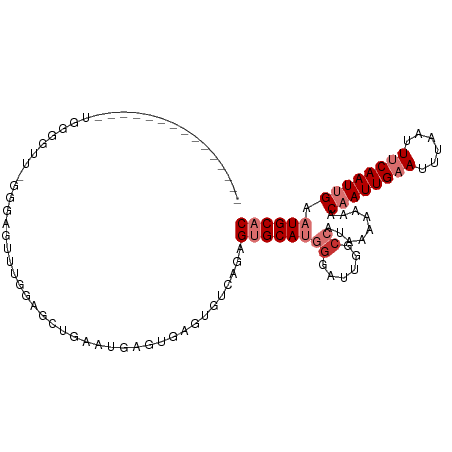
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:04 2011