| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,142,408 – 7,142,501 |
| Length | 93 |
| Max. P | 0.935930 |

| Location | 7,142,408 – 7,142,501 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.68655 |
| G+C content | 0.44984 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -9.96 |
| Energy contribution | -9.93 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
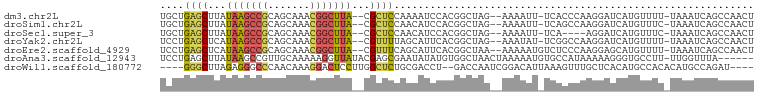
>dm3.chr2L 7142408 93 + 23011544 UGCUGAGCUUAUAAGCCGCAGCAAACGGCUUA--CGCUCCAAAAUCCACGGCUAG--AAAAUU-UCACCCAAGGAUCAUGUUUU-UAAAUCAGCCAACU .(((((((...(((((((.......)))))))--.))..((..((((..((...(--(.....-))..))..))))..))....-....)))))..... ( -20.80, z-score = -1.36, R) >droSim1.chr2L 6931978 93 + 22036055 UGCUGAGCUUAUAAGCCGCAGCAAACGGCUUA--CGCUCCAACAUCCACGGCUAG--AAAAUU-UCAGCCAAGGAUCAUGUUUC-UAAAUCAGCCAACU .(((((((...(((((((.......)))))))--.))......((((..((((.(--(.....-))))))..))))........-....)))))..... ( -26.10, z-score = -2.72, R) >droSec1.super_3 2666068 89 + 7220098 UGCUGAGCUUAUAAGCCGCAGCAAACGGCUUA--CGCUCCAACAUCCACGGCUAG--AAAAUU-UCA----AGGAUCAUGUUUC-UAAAUCAGCCAACU ....((((...(((((((.......)))))))--.))))..........((((.(--(.....-)).----((((......)))-).....)))).... ( -20.70, z-score = -1.45, R) >droYak2.chr2L 16560782 93 - 22324452 UCCUGAGCUCAUAAGCCGCAGCAAACGGCUUA--CGUUUUAGCAUUCACGGCUAG--AAAUAU-UCGGCCAAGGAUCAUGUUUU-UAAAUCAGCCAACU ((((..(((..(((((((.......)))))))--..(((((((.......)))))--))....-..)))..)))).........-.............. ( -22.30, z-score = -1.41, R) >droEre2.scaffold_4929 16057775 94 + 26641161 UCCUGAGCUCAUAAGCCGCAGCAAACGGCUUA--CGUUUCAGCAUUCACGGCUAA--AAAAAUGUCUCCCAAGGAGCAUGUUUU-UAAAUCAGCCAACU ..(((((..(.(((((((.......)))))))--.).))))).......((((((--(((.(((.((((...))))))).))))-).....)))).... ( -25.90, z-score = -2.97, R) >droAna3.scaffold_12943 992399 92 - 5039921 UCCUGAGCUUAUAAGCCGUUGCAAAAAGGUUAUACGAGCGAAUAUAUGUGGCUAACUAAAAAUGUGCCAUAAAAAGGGUGCCUU-UUGGUUUA------ ((((..(((..((((((..........)))).))..))).......((((((.............))))))...)))).(((..-..)))...------ ( -20.12, z-score = -0.22, R) >droWil1.scaffold_180772 3214292 89 - 8906247 ----GGGCUUAGAGGGCCCAACAAAGGACUCCUUGGCUCUGCGACCU--GACCAAUCGGACAUUAAAGUUUGCUCACAUGCCACACAUGCCAGAU---- ----(((((.....)))))......((......((((...((((.((--..((....)).......)).))))......))))......))....---- ( -19.80, z-score = 0.83, R) >consensus UCCUGAGCUUAUAAGCCGCAGCAAACGGCUUA__CGCUCCAACAUCCACGGCUAG__AAAAAU_UCACCCAAGGAUCAUGUUUU_UAAAUCAGCCAACU ....((((...(((((((.......)))))))...))))............................................................ ( -9.96 = -9.93 + -0.04)
| Location | 7,142,408 – 7,142,501 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.68655 |
| G+C content | 0.44984 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.23 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
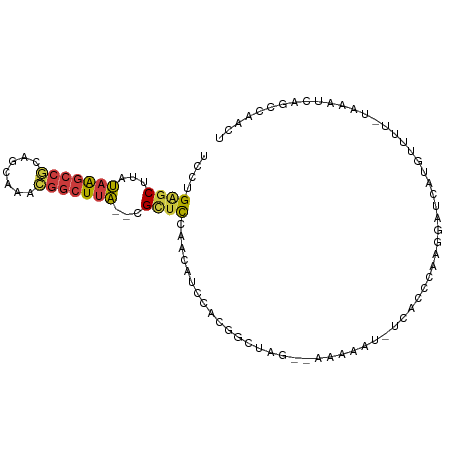
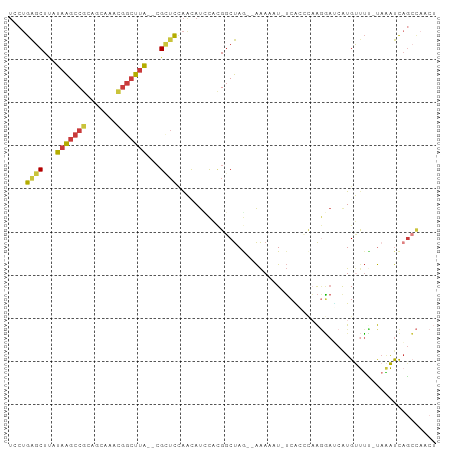
>dm3.chr2L 7142408 93 - 23011544 AGUUGGCUGAUUUA-AAAACAUGAUCCUUGGGUGA-AAUUUU--CUAGCCGUGGAUUUUGGAGCG--UAAGCCGUUUGCUGCGGCUUAUAAGCUCAGCA .....(((......-....((.(((((.(((.((.-......--.)).))).))))).))(((((--((((((((.....)))))))))..))))))). ( -32.70, z-score = -2.72, R) >droSim1.chr2L 6931978 93 - 22036055 AGUUGGCUGAUUUA-GAAACAUGAUCCUUGGCUGA-AAUUUU--CUAGCCGUGGAUGUUGGAGCG--UAAGCCGUUUGCUGCGGCUUAUAAGCUCAGCA .....(((......-....((..((((.((((((.-......--.)))))).))))..))(((((--((((((((.....)))))))))..))))))). ( -35.60, z-score = -3.20, R) >droSec1.super_3 2666068 89 - 7220098 AGUUGGCUGAUUUA-GAAACAUGAUCCU----UGA-AAUUUU--CUAGCCGUGGAUGUUGGAGCG--UAAGCCGUUUGCUGCGGCUUAUAAGCUCAGCA .....(((......-....((..((((.----((.-......--.....)).))))..))(((((--((((((((.....)))))))))..))))))). ( -25.70, z-score = -0.86, R) >droYak2.chr2L 16560782 93 + 22324452 AGUUGGCUGAUUUA-AAAACAUGAUCCUUGGCCGA-AUAUUU--CUAGCCGUGAAUGCUAAAACG--UAAGCCGUUUGCUGCGGCUUAUGAGCUCAGGA ..((((((((((..-.......))))...))))))-......--....((.(((..(((....((--((((((((.....)))))))))))))))))). ( -26.90, z-score = -1.61, R) >droEre2.scaffold_4929 16057775 94 - 26641161 AGUUGGCUGAUUUA-AAAACAUGCUCCUUGGGAGACAUUUUU--UUAGCCGUGAAUGCUGAAACG--UAAGCCGUUUGCUGCGGCUUAUGAGCUCAGGA ....((((((....-((((.(((((((...)))).)))))))--)))))).......((((..((--((((((((.....))))))))))...)))).. ( -31.90, z-score = -2.63, R) >droAna3.scaffold_12943 992399 92 + 5039921 ------UAAACCAA-AAGGCACCCUUUUUAUGGCACAUUUUUAGUUAGCCACAUAUAUUCGCUCGUAUAACCUUUUUGCAACGGCUUAUAAGCUCAGGA ------....((((-((((...........((((.............))))..(((((......))))).))))))......((((....))))..)). ( -12.92, z-score = 0.66, R) >droWil1.scaffold_180772 3214292 89 + 8906247 ----AUCUGGCAUGUGUGGCAUGUGAGCAAACUUUAAUGUCCGAUUGGUC--AGGUCGCAGAGCCAAGGAGUCCUUUGUUGGGCCCUCUAAGCCC---- ----....(((.....((((.(((((.(..(((.............))).--.).)))))..))))(((.((((......)))))))....))).---- ( -27.12, z-score = -0.28, R) >consensus AGUUGGCUGAUUUA_AAAACAUGAUCCUUGGCUGA_AUUUUU__CUAGCCGUGGAUGUUGGAGCG__UAAGCCGUUUGCUGCGGCUUAUAAGCUCAGCA ....(((((....................................))))).................((((((((.....))))))))........... ( -9.10 = -9.23 + 0.13)

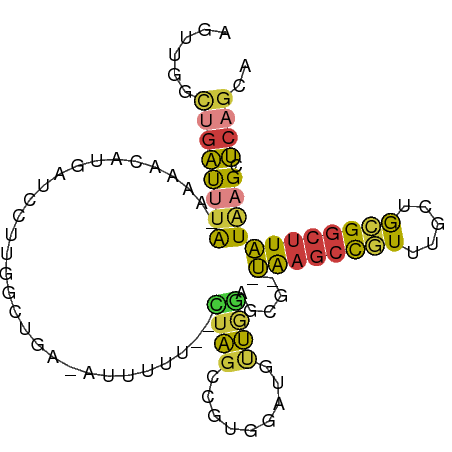
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:22:59 2011