| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,778,802 – 7,778,894 |
| Length | 92 |
| Max. P | 0.996033 |

| Location | 7,778,802 – 7,778,894 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 66.29 |
| Shannon entropy | 0.73525 |
| G+C content | 0.47320 |
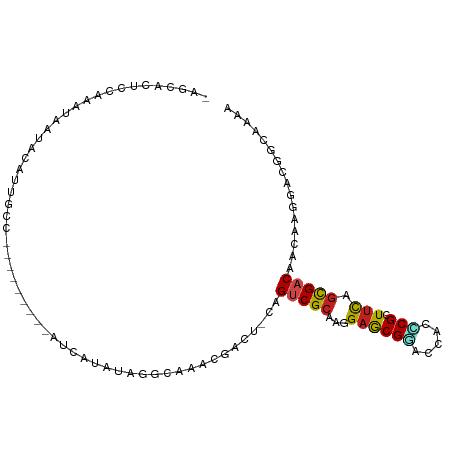
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -10.54 |
| Energy contribution | -10.22 |
| Covariance contribution | -0.31 |
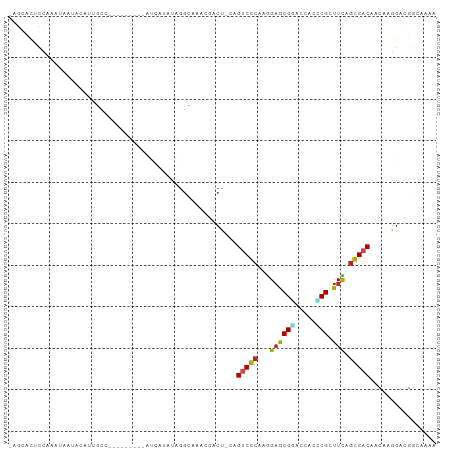
| Combinations/Pair | 1.55 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.996033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
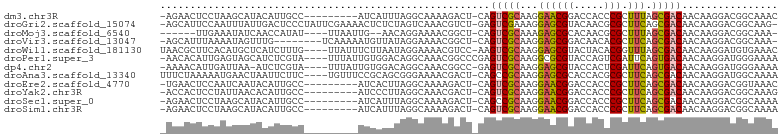
>dm3.chr3R 7778802 92 + 27905053 -AGAACUCCUAAGCAUACAUUGCC---------AUCAUUUAGGCAAAAGACU-CAGUCGCAAGGAACGGACCACCCGCUUUAGCGACAACAAGGACGGCAAAC -.(...((((.........(((((---------........)))))......-..(((((((((..(((.....))))))).)))))....))))...).... ( -23.20, z-score = -2.70, R) >droGri2.scaffold_15074 1667931 100 - 7742996 -AGCAUUCCAAUUUAUUGACUCCCUAUUCGAAAACUCUCUAGUCAAACGUCU-GAGUCGAAAGGAGCGUACAACGCGCUUCAGCGACAACAAGGACGGCAAG- -.((...........((((((...................)))))).(((((-..((((...(((((((.....)))))))..)))).....)))))))...- ( -26.81, z-score = -2.08, R) >droMoj3.scaffold_6540 5705891 89 + 34148556 ------UUGAAAUAUCAACCAUAU----UUAAUUG--AACAGGAAAACGGCU-CAGUCGCAAAGAGCGCACAACGCGCUUUAGCGACAACAAGGACGGCAAA- ------((((....))))((....----.....((--(.(.(.....).).)-))(((((..(((((((.....))))))).))))).....))........- ( -20.80, z-score = -2.22, R) >droVir3.scaffold_13047 1687881 92 + 19223366 -AGCAUUUAAAAUAGUUUG--------UCAAAAAUGUUAUAGGAAAACGGCU-CAGUCGCAAGGAGCGGACAACACGCUUCAGCGACAACAAGGACGGCAAA- -..............((((--------((.....((((.......))))...-..(((((..((((((.......)))))).))))).........))))))- ( -21.80, z-score = -2.15, R) >droWil1.scaffold_181130 3276098 98 + 16660200 UAACGCUUCACAUGCUCAUCUUUG----UUAUUUCUUAAUAGGAAAACGUCC-AAGUCGCAAGGAGCGUACUACACGGUUUAGCGACAACAAGGAUGUGAAAC ....((.......)).((((((((----((...........(((.....)))-..(((((..(((.(((.....))).))).)))))))))))))))...... ( -25.50, z-score = -2.28, R) >droPer1.super_3 5314908 98 + 7375914 -AACACAUUGAGUAGCAUCUCGUA----UUUAUUGUGGACAGGCAAACGGCCCGAGUCGCAAGGCGCGUACCAGUCGAUUCAGUGACAACAAGGAUGGGAAAA -................((((((.----....((((((...(((.....)))((.(((....))).))..)).((((......)))).))))..))))))... ( -22.90, z-score = 0.30, R) >dp4.chr2 22536022 96 + 30794189 -AAAACAUUGAUUAA-AUCUCGUA----UUUAUUGUGGACAGGCAAACGGCC-GAGUCGCAAGGAGCGUACCACUCGAUUCAGUGACAACAAGGAUGGGAAAA -..............-.((((((.----(((.((((.(.(.(((.....)))-((((((...((......))...)))))).)).)))).))).))))))... ( -21.70, z-score = -0.63, R) >droAna3.scaffold_13340 8262553 98 - 23697760 UUUCUAAAAAUGAACUAAUUCUUC----UGUUUCCGCAGCGGGAAAACGACU-CAGCCGCAAGGAGCGCACCACGCGCUUCAGCGACAACAAGGAUGGCAAAA .................(((((..----((((..(((.((((..........-...))))..(((((((.....))))))).)))..)))))))))....... ( -26.72, z-score = -1.23, R) >droEre2.scaffold_4770 3905109 92 - 17746568 -UGAACUCCAAUCAAUACAUUGCC---------AUCACUUAGGCAAAAGACU-CAGUCGCAAGGAACGGACCACCCGCUUCAGCGACAACAAGGACGGUAAAC -.....(((..........(((((---------........)))))......-..(((((...((((((.....))).))).))))).....)))........ ( -23.30, z-score = -2.85, R) >droYak2.chr3R 12191458 92 - 28832112 -ACCACUCCUAUUAACACAUUGCC---------AUCCCUUAGGCAAACGACU-CAGUCGCAAGGAACGGACCACCCGCUUCAGCGACAACAAGGACGGCAAAG -.((..((((.........(((((---------........)))))......-..(((((...((((((.....))).))).)))))....)))).))..... ( -25.10, z-score = -3.51, R) >droSec1.super_0 6929822 92 - 21120651 -AGAACUCCUAAGCAUACAUUGCC---------AUCAUUUAGGCAAAAGACU-CAGCCGCAAGGAACGGACCACCCGCUUCAGCGACAACAAGGACGGCAAAA -..................(((((---------.((.....(((........-..)))((...((((((.....))).))).)).........)).))))).. ( -19.40, z-score = -0.98, R) >droSim1.chr3R 13903122 92 - 27517382 -AGAACUCCUAAGCAUACAUUGCC---------AUCAUUUAGGCAAAAGACU-CAGUCGCAAGGAACGGACCACCCGCUUCAGCGACAACAAGGACGGCAAAA -.(...((((.........(((((---------........)))))......-..(((((...((((((.....))).))).)))))....))))...).... ( -24.20, z-score = -2.80, R) >consensus _AGCACUCCAAAUAAUACAUUGCC_________AUCAUAUAGGCAAACGACU_CAGUCGCAAGGAGCGGACCACCCGCUUCAGCGACAACAAGGACGGCAAAA .......................................................(((((...((((((.....))).))).)))))................ (-10.54 = -10.22 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:08:01 2011