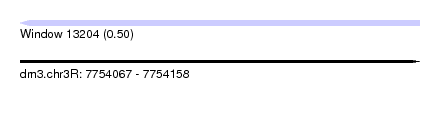
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,754,067 – 7,754,158 |
| Length | 91 |
| Max. P | 0.500000 |

| Location | 7,754,067 – 7,754,158 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.35 |
| Shannon entropy | 0.46021 |
| G+C content | 0.46097 |
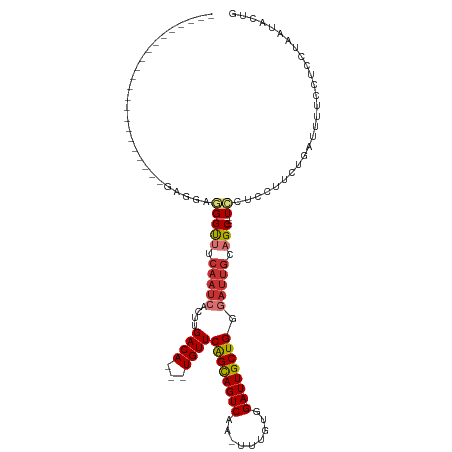
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -13.44 |
| Energy contribution | -12.96 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
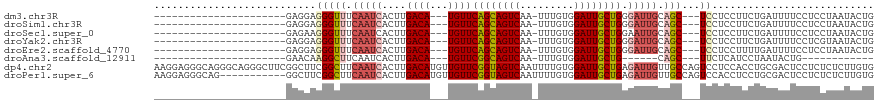
Download alignment: ClustalW | MAF
>dm3.chr3R 7754067 91 - 27905053 ----------------------GAGGAGGGUUUCAAUCACUUGACA---UGUUCAGCAGUCAA-UUUGUGGAUUGCUGGGAUUGCAGC---UCCUCCUUCUGAUUUUCCUCCUAAUACUG ----------------------((((((((((.(((((((......---.))(((((((((..-......)))))))))))))).)))---.)))))))..................... ( -29.30, z-score = -2.22, R) >droSim1.chr3R 13876945 91 + 27517382 ----------------------GAGGAGGGUUUCAAUCACUUGACA---UGUUCAGCAGUCAA-UUUGUGGAUUGCUGGGAUUGCAGC---UCCUCCUUCUGAUUUUCCUCCUAAUACUG ----------------------((((((((((.(((((((......---.))(((((((((..-......)))))))))))))).)))---.)))))))..................... ( -29.30, z-score = -2.22, R) >droSec1.super_0 6905045 91 + 21120651 ----------------------GAGAAGGGUUUCAAUCACUUGACA---UGUUCAGCAGUCAA-UUUGUGGAUUGCUGGAAUUGCAGC---UCCUCCUUCUGAUUUUCCUCCUAAUACUG ----------------------.(((((((..((((....))))((---..((((((((((..-......))))))))))..))....---...)))))))................... ( -22.30, z-score = -0.75, R) >droYak2.chr3R 12166414 91 + 28832112 ----------------------GAGGAGGGUUUCAAUCACUUGACA---UGUUCAGCAGUCAA-UUUGUGGAUUGCUGGGAUUGCAGC---UCCUCCUUCUGAUUUUCCUCGUAAUACUG ----------------------((((((((((.(((((((......---.))(((((((((..-......)))))))))))))).)))---.)))))))..................... ( -29.30, z-score = -2.27, R) >droEre2.scaffold_4770 3880130 91 + 17746568 ----------------------GAGGAGGGUUUCAAUCACUUGACA---UGUUCAGCAGUCAA-UUUGUGGAUUGCUGGGAUUGCAGC---UCCUCCUUUUGAUUUUCCUCCUAAUACUG ----------------------((((((((((.(((((((......---.))(((((((((..-......)))))))))))))).)))---.)))))))..................... ( -29.30, z-score = -2.42, R) >droAna3.scaffold_12911 2421356 73 - 5364042 ----------------------GAACAAGGCUUCAAUCACUUGACA---UGUUCGGCAGUCAA-UUUGUGGAUUGCUG------CAGC---UUCUCAUCCUAAUACUG------------ ----------------------...((((..........))))...---.(((((((((((..-......))))))))------.)))---.................------------ ( -14.70, z-score = -0.23, R) >dp4.chr2 5439665 120 + 30794189 AAGGAGGGCAGGGCAGGGCUUCGGCUUCGGCUUCAAUCACUUGACAUGUUGUUCGGUAGUCAAUUUUGUGGAUUGCUGAGAUUGUUGCCAGUCCUCCACCUGCGACUCCUCUCUCUUGUG .(((((.((((((.(((((....))...(((..(((((.....((.....))(((((((((.........))))))))))))))..)))...))).).)))))..))))).......... ( -43.90, z-score = -1.80, R) >droPer1.super_6 5456496 109 + 6141320 AAGGAGGGCAG-----------GGCUUCGGCUUCAAUCACUUGACAUGUUGUUCGGUAGUCAAUUUUGUGGAUUGCUGAGAUUGUUGCCAGUCCACCUCCUGCGACUCCUCUCUCUUGUG .((((((...(-----------((((..(((..(((((.....((.....))(((((((((.........))))))))))))))..)))))))).))))))................... ( -35.80, z-score = -1.34, R) >consensus ______________________GAGGAGGGUUUCAAUCACUUGACA___UGUUCAGCAGUCAA_UUUGUGGAUUGCUGGGAUUGCAGC___UCCUCCUUCUGAUUUUCCUCCUAAUACUG ......................(((..(((..((((....)))).......((((((((((.........))))))))))............)))..))).................... (-13.44 = -12.96 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:58 2011