
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,752,165 – 7,752,266 |
| Length | 101 |
| Max. P | 0.624368 |

| Location | 7,752,165 – 7,752,266 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.11 |
| Shannon entropy | 0.46190 |
| G+C content | 0.42616 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.59 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
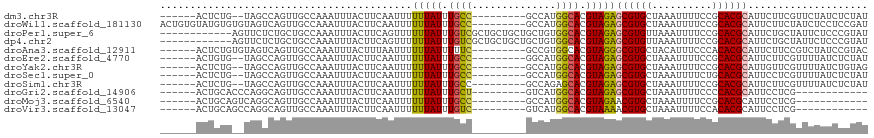
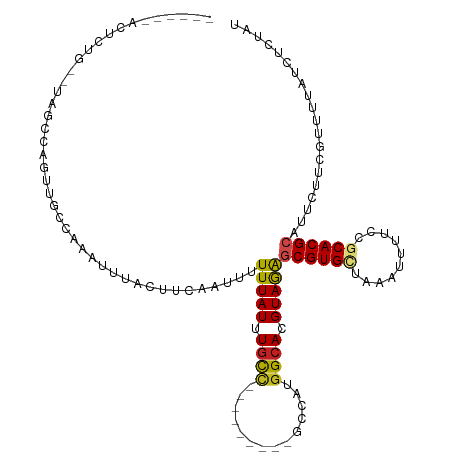
>dm3.chr3R 7752165 101 - 27905053 ------ACUCUG--UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAUGGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUCUUCGUUCUAUCUCUAU ------......--..((((...(((((((...............)))))..---------))..))))(((.(((((((((..........))))))..))).)))........... ( -21.26, z-score = -2.11, R) >droWil1.scaffold_181130 15695730 109 - 16660200 ACUGUGUAUGUGUGUAGUCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAUGGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUCUUCUAUCUCCUCCGAU .....(.(((((((..(..(((((((((((...............)))))..---------))..(((((((.....)))))))))))..)..))))))).)................ ( -21.76, z-score = -0.88, R) >droPer1.super_6 5454297 106 + 6141320 ------------AGUUCUCUGCUGCCAAAUUUACUUCAGUUUUUUAUUUGUCGCUGCUGCUGCUGUGGCACGUAGAGCGUGUUAAAUUUUCCGCACGCAUUCUGCUAUUCUCCCGUAU ------------.....((((((((((......(..((((..(......)..))))..)......))))).)))))((((((..........)))))).................... ( -23.50, z-score = -0.72, R) >dp4.chr2 5437524 106 + 30794189 ------------AGUUCUCUGCUGCCAAAUUUACUUCAGUUUUUUAUUUGUCGCUGCUGCUGCUGUGGCACGUAGAGCGUGUUAAAUUUUCCGCACGCAUUCUGCUAUUCUCCCGUAU ------------.....((((((((((......(..((((..(......)..))))..)......))))).)))))((((((..........)))))).................... ( -23.50, z-score = -0.72, R) >droAna3.scaffold_12911 2419541 103 - 5364042 ------ACUCUGUGUAGUCAGUUGCCAAAUUUACUUUAAUUUUUUAUUUUUC---------GCCGUGGCACGUAGGGCGUGCUACAUUUCCCACACGCAUUCUUCCGUCUAUCCGUAC ------....((((((((.((((....)))).))).................---------...((((((((.....)))))))).........)))))................... ( -19.40, z-score = -1.04, R) >droEre2.scaffold_4770 3878250 101 + 17746568 ------ACUGUG--UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GGCAUGGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUCUUCGUUUUAUCUCUAU ------.(((((--(.((((((((.(((((...............))))).)---------))).)))))))))).((((((..........)))))).................... ( -25.56, z-score = -2.56, R) >droYak2.chr3R 12164485 101 + 28832112 ------ACUCUG--UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAUGGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUGUUCGUUUUAUCUGUAG ------.((.((--(.((((...(((((((...............)))))..---------))..))))))).)).((((((..........)))))).................... ( -20.56, z-score = -0.85, R) >droSec1.super_0 6903171 101 + 21120651 ------ACUCUG--UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAUGGCACGUAGAGCGUGCUAAAUUUUCUGCACGCAUUCCUCGUUUUAUCUCUAU ------.((.((--(.((((...(((((((...............)))))..---------))..))))))).)).((((((..........)))))).................... ( -21.16, z-score = -1.96, R) >droSim1.chr3R 13875083 101 + 27517382 ------ACUCUG--UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAGAGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUCUUCGUUUUAUCUCUAU ------.(((((--..((.(((....(((((......)))))...))).)).---------.).)))).(((.(((((((((..........))))))..))).)))........... ( -20.10, z-score = -1.75, R) >droGri2.scaffold_14906 466661 91 - 14172833 ------ACUGCACCCAGGCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCU---------GUCAUGGCACGUAGAGCGUGCUAAAUUUUCCCCACGCAUUCCUCG------------ ------..(((.....((((((....(((((......))))).......)))---------))).(((((((.....)))))))............))).......------------ ( -19.80, z-score = -1.68, R) >droMoj3.scaffold_6540 20815025 91 - 34148556 ------ACUGCAGUCAGGCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC---------GCCAUGGCACGUAGAACGUGCUAAAUUUUCCGCACGCAUUCCUCG------------ ------(((((......)))))(((................((((((.((((---------.....)))).)))))).((((..........))))))).......------------ ( -20.60, z-score = -1.34, R) >droVir3.scaffold_13047 3812771 91 - 19223366 ------ACUGCAGCCAGGCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGUC---------GUCAUGGCACGUAAAACGUGCUAAAUUUUCCACACGCAUUCCUCG------------ ------..(((.....(((....)))..........................---------....(((((((.....)))))))............))).......------------ ( -18.00, z-score = -1.46, R) >consensus ______ACUCUG__UAGCCAGUUGCCAAAUUUACUUCAAUUUUUUAUUUGCC_________GCCAUGGCACGUAGAGCGUGCUAAAUUUUCCGCACGCAUUCUUCGUUUUAUCUCUAU ..........................................(((((.((((..............)))).)))))((((((..........)))))).................... (-11.55 = -11.59 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:57 2011