| Sequence ID | dm3.chr3R |
|---|---|
| Location | 7,751,433 – 7,751,525 |
| Length | 92 |
| Max. P | 0.797691 |

| Location | 7,751,433 – 7,751,525 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Shannon entropy | 0.37409 |
| G+C content | 0.40650 |
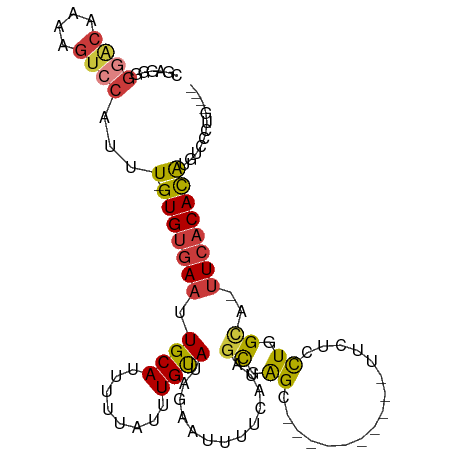
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -13.14 |
| Energy contribution | -14.14 |
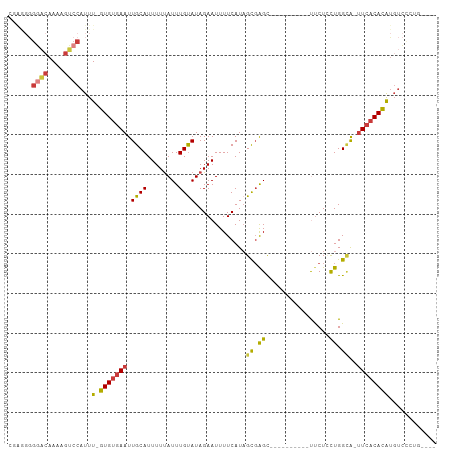
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
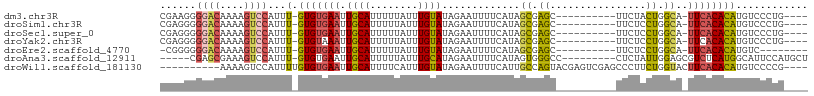
>dm3.chr3R 7751433 92 + 27905053 CGAAGGGGACAAAAGUCCAUUU-GUGUGAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC----------UUCUACUGGCA-UUCACACAUGUCCCUG---- ....(((((((..........(-((((((((((((........)))).............((.((.----------.....)).)))-))))))))))))))).---- ( -25.90, z-score = -2.73, R) >droSim1.chr3R 13874344 92 - 27517382 CGAGGGGGACAAAAGUCCAUUU-GUGUGAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC----------UUCUCCUGGCA-UUCACACAUGUCCCUG---- ..((((((((....))))...(-((((((((.((..((((((....))))))......(((.(((.----------..)))))))))-))))))))...)))).---- ( -28.50, z-score = -3.09, R) >droSec1.super_0 6902436 92 - 21120651 CGAGGGGGACAAAAGUCCAUUU-GUGUGAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC----------UUCUCCUGGCA-UUCACACAUGUCCCUG---- ..((((((((....))))...(-((((((((.((..((((((....))))))......(((.(((.----------..)))))))))-))))))))...)))).---- ( -28.50, z-score = -3.09, R) >droYak2.chr3R 12163661 92 - 28832112 CGAGGGGGACAAAAGUCCAUUU-GUGUAAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC----------UUCUCCUGGCA-UUCACACAUGUCCCUG---- ..((((((((....))))...(-((((....(((..((((((....))))))......(((.(((.----------..)))))))))-...)))))...)))).---- ( -22.40, z-score = -1.47, R) >droEre2.scaffold_4770 3877516 87 - 17746568 -CGGGGGGACAAAAGUCCAUUU-GUGUGAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC----------UUCUCCUGGCA-UUCACACAUGUC-------- -.....((((....))))...(-((((((((.((..((((((....))))))......(((.(((.----------..)))))))))-))))))))....-------- ( -22.40, z-score = -1.77, R) >droAna3.scaffold_12911 2419020 93 + 5364042 -----CGAGCGAAAGUCCAUUU-GUGUGAAUUGCAUUUUUAUUUGCAUAGAAUUUUCAUAGUGGGCC---------CUCUAUUGGAGCGUCUCAUGGCAUUCCAUGCU -----..(((((..(((((((.-(((.((((((((........))))....)))).)))))))))).---------.))...(((((.(((....))).))))).))) ( -25.40, z-score = -1.30, R) >droWil1.scaffold_181130 15695376 94 + 16660200 ----------AAAAGUCCAUUUUGUGUGAAUUGCAUUUUCAUUUGUAUAGAAUUUUCAUUGCCAGUACGAGUCGAGCCCUUCUGGUACUUCACACAUGUCCCCG---- ----------............((((((((.((((........))))............((((((...(((.......))))))))).))))))))........---- ( -17.30, z-score = -1.14, R) >consensus CGAGGGGGACAAAAGUCCAUUU_GUGUGAAUUGCAUUUUUAUUUGUAUAGAAUUUUCAUAGCGAGC__________UUCUCCUGGCA_UUCACACAUGUCCCUG____ ......((((....)))).....(((((((.(((..((((((....))))))..(((.....)))...................))).)))))))............. (-13.14 = -14.14 + 1.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:07:57 2011